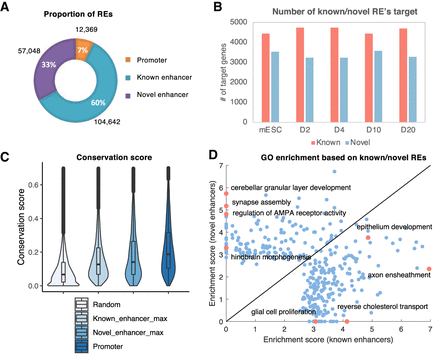
Identification and annotation of novel enhancers. (A) Pie charts of the promoter, known enhancers, and novel enhancers in REs. (B) Bar plot of numbers of known enhancers and novel enhancers at different time points. (C) Conservation score distribution of different sets of REs. (D) Comparison of GO enrichment on known enhancers’ targets and novel enhancers’ targets on D10. Top 500 specifically expressed genes in each group are chosen to perform GO analysis. The x-axis represents enrichment score on known enhancers’ targets, and the y-axis represents that on novel enhancers’ targets. Each dot represents one GO term. The enrichment score is defined as the geometric mean of fold change and −log10 P-value.











