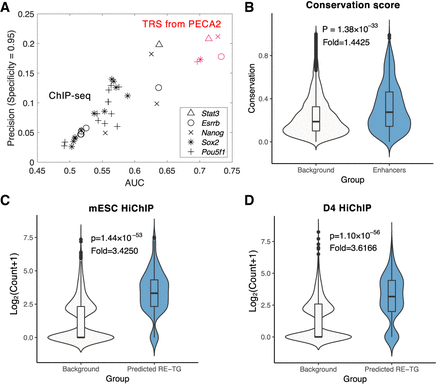
PECA2 infers accurate gene regulation supported by ChIP-seq, shRNA knockdown, and HiChIP experiments. (A) Comparison of PECA2 TRS with ChIP-seq experiment on five important regulators in mESC by taking the knockdown data as ground truth. Shapes represent different transcription factors and colors represent different methods. Red represents results from PECA2 and black represents results from ChIP-seq experiment. (B) Conservation score distribution comparison between REs predicted to regulate at least one gene and randomly selected REs. (C,D) Validation of PECA2 predicted RE-TG pairs by the HiChIP experiment on mESC and RA D4. Background RE-TG pairs are randomly selected to have the same distance distribution as the predicted RE-TG pairs. “Fold” represents fold change of average read count of predicted RE-TG pairs versus background RE-TG pairs.











