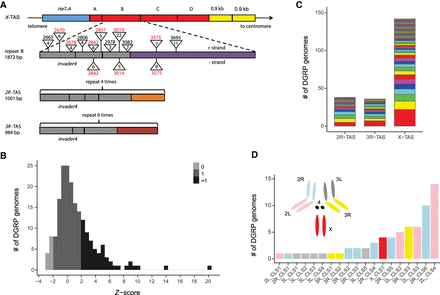
P-element insertions in TAS and non-TAS piRNA clusters. (A) The structure of TAS arrays (modeled after Asif-Laidin et al. 2017). X-TAS contains four tandem repeats (A–D in red) located between a HeT-A retrotransposon array and two 0.9-kb repeats (Karpen and Spradling 1992). Repeat A is degenerated. Repeats B–D are ∼1.8 kb in length and are highly similar to each other (>95% identity). Repeat B is compared to repeats from 2R and 3R-TAS. Each repeat of 2R, 3R, and X-TAS contains several invader4 retrotransposon long terminal repeats (LTRs) (gray) (Bergman et al. 2006), as well as other short homologous fragments (41–131 bp), with 909 bp being unique to the X-TAS repeat (Asif-Laidin et al. 2017). P-element insertions located in X-TAS and present in more than two DGRP genomes are depicted as triangles, with the numeric value above corresponding to the breakpoint, and the numeric value inside corresponding to the number of DGRP genomes containing the insertion. Breakpoints in red correspond to insertion hotspots (Karpen and Spradling 1992). (B) The distribution of Z-scores for P/TAS-derived read counts among DGRP libraries. DGRP genomes with Z < −1.96, −1.96 < Z < 1.96, and Z > 1.96 were estimated to have 0, 1, and >1 P-element, respectively. (C) Multiple P-element insertion sites were detected in 2R, 3R, and X-TAS. Each color represents a unique P-element insertion. (D) The frequency of P-element insertions in non-TAS ancestral piRNA clusters (based on our annotation set of 159 piRNA clusters) across all major chromosome arms.











