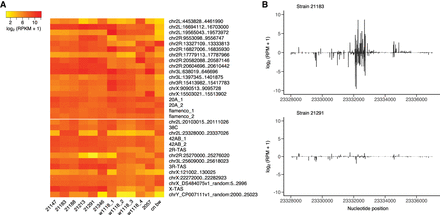
piRNA cluster annotation in P-element free strains. (A) Activity of 32 piRNA clusters in ancestral (P-element free) strains of D. melanogaster. Each column represents a small-RNA sequencing library (biological replicates are combined), and each row represents a piRNA cluster annotated in at least one of these libraries. Coordinates of piRNA clusters are based on the D. melanogaster release 6 assembly (dm6) (Hoskins et al. 2015). piRNA cluster expression levels are estimated by reads per kilo base per million mapped reads (RPKM) and transformed to log2 scale [log2 (RPKM + 1)]. Clusters above the white line are unistrand piRNA clusters, and those below the white line are dual-strand piRNA clusters. Details on small-RNA library prep, which may be related to differences in annotated piRNA clusters between libraries from w1118, are provided in Supplemental Table S1. (B) An example of a polymorphism in piRNA cluster activity in an annotated cluster on Chromosome 2L (23,328,000–23,337,026). Abundant piRNAs are detected from strain 21183, whereas strain 21291 produces few piRNAs. Only uniquely mapping piRNAs are considered. piRNA density is measured in reads per million mapped reads (RPM) and transformed to log2 scale [log2 (RPM + 1)]. Positive values represent piRNAs mapped to sense strand of the reference genome, and negative values represent piRNAs from antisense strand.











