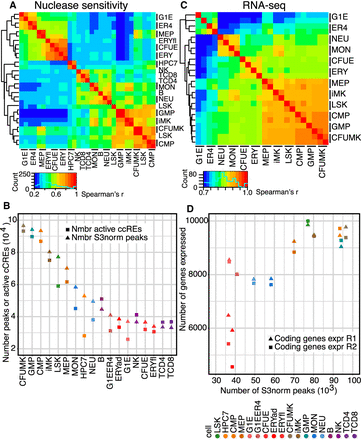
Global comparisons of nuclease accessibility profiles and transcriptomes across mouse hematopoietic cell types. (A) Heatmap of the hierarchical clustering of nuclease-sensitive elements (ATAC-seq and DNase-seq, using S3norm for normalization), with Spearman's rank correlation r as the similarity measure, and 1 − r as the distance measure for hierarchical clustering across 18 cell types. Results include replicates for cell types with replicated data (indicated by bars next to the cell type name). (B) Numbers of dynamic cCREs in each cell type, determined from ATAC-seq and DNase-seq profiles and analyzed as both peak calls from HOMER (Heinz et al. 2010) or from peaks after S3 normalization. (C) Heatmap of the hierarchical clustering of RNA-seq (TPM values for all genes, quantile normalized, showing replicates), with Spearman's r as the similarity measure. (D) Concordant decreases during hematopoietic differentiation in nuclease accessibility and expressed genes, shown as the association between numbers of genes expressed and numbers of dynamic cCREs across cell populations and types.











