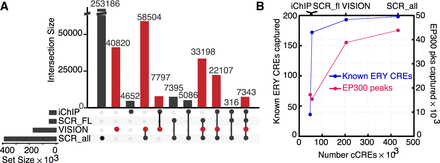
Comparative analysis of VISION cCREs. (A) Overlaps of the VISION cCREs with three other cCRE catalogs. The overlapping cCREs in all four data sets were merged. The numbers of merged cCREs in each set are labeled on each row, and the numbers in each level of overlap are shown in columns, visualized using an UpSet plot (Lex et al. 2014). The sets compared with the VISION cCREs were the Blood Enhancer Catalog derived from iChIP data (iChIP) (Lara-Astiaso et al. 2014), the SCREEN cCREs specific to mouse fetal liver at E14.5 (SCR_FL), and those for all tissues and cell types in mouse (SCR_all). (B) The VISION cCREs capture known regulatory elements and orthogonal predicted cCREs. The number of known CREs that are also present in each cCRE collection was plotted against the number of regulatory elements (known or inferred) in each data set. The EP300 peaks were deduced from EP300 ChIP-seq data from ENCODE, reprocessed by VISION pipelines, from FL E14.5, MEL, and CH12 cells. Replicated peaks were combined into one data set and merged to get more than 60,000 peaks. The number of known EP300 peaks that were also present in each cCRE collection was plotted against the number of cCREs in each data set.











