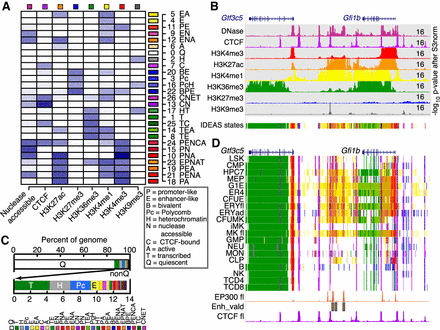
Segmentation of the epigenomes of hematopoietic cells after integrative modeling with IDEAS. (A) Heatmap of the emission frequencies of each of the 27 states discovered by IDEAS, with state number and function-associated labels. Each letter in the label indicates a function associated with the combination of features in each state, defined in the box. The indicator for transcribed is H3K36me3, active is H3K27ac, enhancer-like is H3K4me1>H3K4me3, promoter-like is H3K4me3>H3K4me1, heterochromatin is H3K9me3, and polycomb is H3K27me3. (B) Example of normalized epigenetic data from ERY in fetal liver around the Gfi1b locus, covering 70 kb from Chr 2: 28,565,001–28,635,000 in GRCm38/mm10, used as input to IDEAS for segmentation. The signal tracks are colored distinctively for each feature, with the inferred epigenetic states shown on the last track. The upper limit for signal in each normalized track is given at the right. (C) Bar graphs of the average coverage of genomes by each state. The top graph emphasizes the high abundance of state Q, and the second graph shows the abundances of the 26 nonquiescent states. The key for annotated colors is the same order as the states in the bar graph. (D) Segmentation pattern across cell types around the Gfi1b exemplar locus. Signal tracks for EP300 (ENCSR982LJQ, ENCODE consortium) and CTCF from mouse fetal liver were included for validation and confirmation, along with the locations of enhancers shown to be active (Enh_vald) (Moignard et al. 2013).











