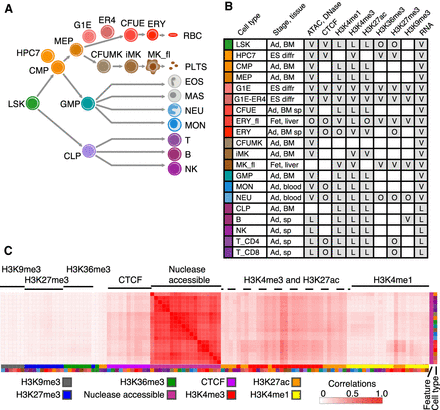
Hematopoietic cell types and data sets used for integrative analysis. (A) Schematic representation of the main lineage commitment steps in hematopoiesis, along with three immortalized cell lines (HPC7, G1E, G1E-ER4) and their approximate position relative to the primary cell populations shown. Abbreviations for cell populations are as follows: (LSK) Lin−Sca1+Kit+, which includes hematopoietic stem cells and multipotent progenitor cells; (CMP) common myeloid progenitor cells; (GMP) granulocyte monocyte progenitor cells; (MEP) megakaryocyte erythrocyte progenitor cells; (CLP) common lymphoid progenitor cells; (CFUE) colony forming unit erythroid; (ERY) erythroblasts; (RBC) red blood cells; (CFUMK) colony forming unit megakaryocyte; (iMK) immature megakaryocytes; (MK_fl) maturing megakaryocytes from fetal liver; (PLTS) platelets; (EOS) eosinophils; (MAS) mast cells; (NEU) neutrophils; (MON) monocytes; (T_CD8) CD8+ T cells; (T_CD4) CD4+ T cells; (B) B cells; (NK) natural killer cells. (B) Available hematopoietic data sets. Shown in each row: cell type along with its representative color, tissue stage ([Ad] adult; [ES diff] embryonic stem cell derived, differentiated), and source ([BM] bone marrow; [sp] spleen, liver, blood). Shaded boxes indicate the presence of the data set, and letters denote the source ([V] VISION; [L] Lara-Astiaso et al. (2014); [O] other). For more information, see Supplemental Table S1. (C) Correlations of nuclease-accessible signals with all features (S3norm normalized) and across cell types. The genome-wide Pearson correlation coefficients r were computed for each cell type–feature pair and displayed as a heatmap after hierarchical clustering (using 1 − r as the distance measure). The features are indicated by a characteristic color (first column on right), and the cell types are indicated in the second column to the right using the same colors as panel B. The full correlation matrix of all features across all cell types is in Supplemental Figure S4.











