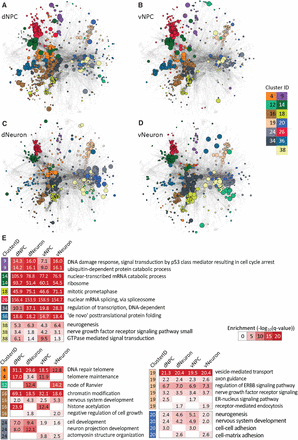
Cell type–specific network clusters of CLIP- and RNA-seq integrative network analysis. (A–D) Cell type–specific network clusters for dNPC (A), vNPC (B), dNeuron (C), and vNeuron (D), identified from our integrative network clustering analysis approach using CLIP and RNA-seq data simultaneously. Node colors correspond to a cluster assignment, and the size of the node is proportional to node diffusion values, which assess the network proximity to the CLIP- and RNA-seq hits. Colors correspond to 13 clusters, seven of which are conserved across all cell types (9, 14, 18, 26, 34, 36, 38) and six clusters that exhibit cell type–specific patterns either among the NPC versus neurons or dorsal versus ventral cell types (4, 12, 16, 19, 20, 24). (E) Enriched Gene Ontology (GO) processes in the 13 clusters depicted in A–D. Cluster IDs and color coding are the same as in A–D. Red-white heat maps show significance of enrichment in a cluster from a particular cell type (−log10(FDR), hypergeometric test).











