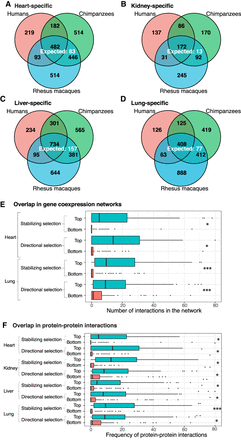
Tissue-specific DE genes (FDR = 0.01). The number of conserved tissue-specific DE genes across all three species is greater than the number expected by chance: (A) heart; (B) kidney; (C) liver; (D) lung. In each tissue, genes with tissue-specific regulatory patterns that are consistent with the action of natural selection (top) are more likely to appear in gene coexpression networks (E) and have an increased number of protein–protein interactions (F) than those that are less consistent with the action of natural selection (bottom). (*) P < 0.05; (***) P < 0.001. The x-axes of E and F are cut off at 80 interactions for readability. In both cases, <5% of the data points are beyond this cutoff.











