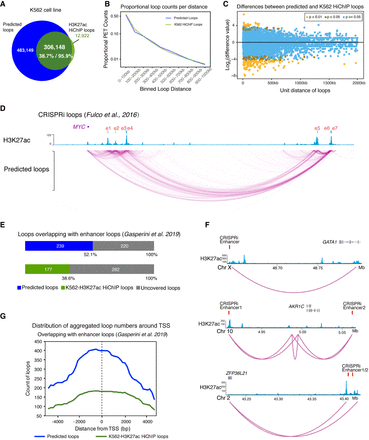
Functional validation of predicted enhancer-mediated interactions. (A) Venn diagram showing the overlap of K562 predicted loops and K562-H3K27ac HiChIP loops. Overall, 306,148 loops were detected by both LoopPredictor and HiChIP experiments, which accounted for 95.9% of the HiChIP loops; 4.1% (12,922 loops) were HiChIP-specific loops. (B) Proportional loop counts per distance for predicted and K562-H3K27ac HiChIP loops. Loops were binned by 100 kb to calculate the proportion. (C) Differences between predicted and K562-H3K27ac HiChIP loops. Loops with a P-value ≥ 0.05 (blue dots) were classified as nonsignificant, loops with a P-value < 0.05 (brown dots) were labeled significant, and differences with a P-value < 0.01 (yellow dots) were marked highly significant. The vast majority of loops showed no significant differences between the two sets of loops. (D) Validation of predicted loops by focused CRISPRi integration (Fulco et al. 2016). Seven previously validated MYC enhancers with strong H3K27ac ChIP-seq signals (blue track) were annotated as e1 through e7 (red track). The predicted loops contacted these published CRISPRi loops. (E) Validation of predicted loops by high-throughput CRISPRi screening integration (Gasperini et al. 2019). A total of 459 high-confidence gene-enhancer loop pairs were overlapped with predicted loops as well as H3K27ac loops. In total, 52% of the high-confidence loop pairs were identified by LoopPredictor. Only 38% of these high-confidence loop pairs were recovered from K562-H3K27ac HiChIP loops. (F) Genome browser tracks for validated enhancer loops identified in E. Validated CRISPRi high-confidence pairs are shown in red; H3K27ac ChIP-seq in blue; predicted loops in purple. (G) Promoter contacts of CRISPRi loops for predicted loops and H3K27ac HiChIP loops. Distribution of aggregated loop numbers by distance of loops from E. The distribution was calculated by ± 4-kb distance from TSS.











