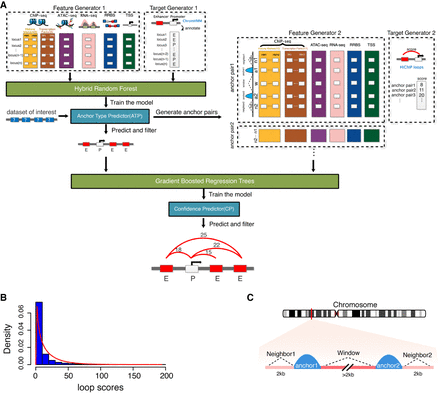
LoopPredictor, an ensemble machine learning model. (A) The LoopPredictor algorithm. H3K27ac and YY1 HiChIP data sets and multi-omics features (e.g., ChIP-seq, RNA-seq, ATAC-seq, and RRBS) were first processed and then integrated to train the model. Targets were defined from the extracted HiChIP anchors via ChromHMM annotation. Next, the trained model and the functional genomics data of interest get put through Anchor type Predictor (ATP), which then identifies the putative topological interactions existing between active genomic regions. The anchor type output from ATP, the newly generated features, and targets of anchor pairs (enhancers and promoters) then get imported into our Confidence Predictor (CP) following Gradient Boosted Regression Trees (GBRTs)-based training. Finally, CP assigns a confidence metric to each predicted chromatin loop, which can be utilized for the filtering of the final LoopPredictor output (Supplemental Methods). (B) Distribution of general HiChIP loop scores after merging four HiChIP data sets (K562-H3K27ac, K562-YY1, HCT116-YY1, and GM12878-H3K27ac). (C) Diagram depicting the loop-associated regions used to gather features in Confidence Predictor.











