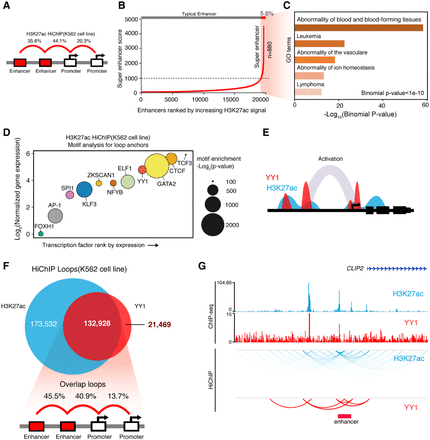
H3K27ac and YY1 HiChIP demarcate active enhancer loops. (A) Proportion of annotated loop types for K562-H3K27ac HiChIP data. Each loop identified with an FDR < 0.05 and pair-end tag number ≥ 2. ChromHMM was used to annotate the anchors, with only enhancer and promoter type anchors being retained. The majority of loops are enhancer-mediated (79.7%). (B) Super-enhancer plot for anchors in K562-H3K27ac HiChIP data. Slope threshold was set at 1000. A total of 880 super-enhancers were found, which accounted for 5.6% of all enhancers. Super-enhancer signal derived from H3K27ac ChIP-seq data. (C) GO analysis for super-enhancer anchors in K562-H3K27ac HiChIP data (binomial P-value < 1 × 10−10). (D) De novo motif enrichment analysis on K562-H3K27ac loops. Transcription factors ranked by normalized gene expression. The size of each point indicates the motif enrichment P-value. The transcription factors with high motif enrichment (–log2[P-value] > 100) and gene expression are shown. The YY1 motif was significantly enriched (−log2[P-value] = 306). (E) Diagram depicting the putative co-enrichment of H3K27ac and YY1. (F) Top: Venn diagram showing the intersect of K562-YY1 and K562-H3K27ac HiChIP data sets. Bottom: Proportion of each annotated loop category from the overlapping H3K37ac and YY1 HiChIP loops (n = 132,928). The majority of the overlapping loops were enhancer-associated (86.4%). (G) Genome browser tracks showing H3K27ac and YY1 ChIP-seq signals and topological interactions.











