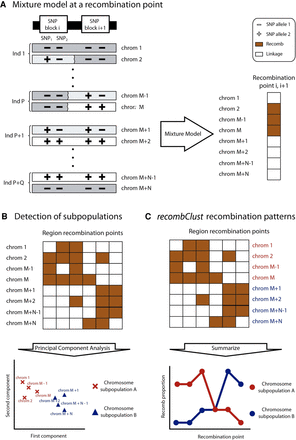
recombClust scheme. recombClust is a method to classify chromosomes into underlying recombination patterns using SNP data, which comprises three steps. (A) Mixture model fitting at a recombination point, flanked by a pair of SNP-blocks. A mixture model classifies the chromosomes into recomb (orange) and linkage (white) groups at a recombination point. For simplicity, SNP-blocks are represented with two alleles. Historical recombination between the SNP-blocks is represented by a broken vertical line between the SNP-blocks. Notice that haplotypes in linkage are those that maximize the likelihood of the mixture model and are not the ancestral haplotypes of the population. (B) Chromosome classification into subpopulations A and B. The mixture models provide a recomb/linkage classification at different recombination points across the target region. In the figure, the orange cells of the classification matrix represent chromosomes classified in the recomb population across recombination points (columns). A principal component (PC) analysis is performed on this matrix. The first PCs reveal clusters of chromosomes for which their recomb/linkage classifications are consistent along the target region and therefore share similar recombination patterns. Each chromosome is assigned to a recombination subpopulation (A: red; B: blue), with chromosomes from 1 to M classified as subpopulation A and chromosomes from M + 1 to M + N as subpopulation B. (C) The recombination pattern for each chromosome subpopulation is reconstructed from the proportion of chromosomes in the recomb group at each point in the genomic region.











