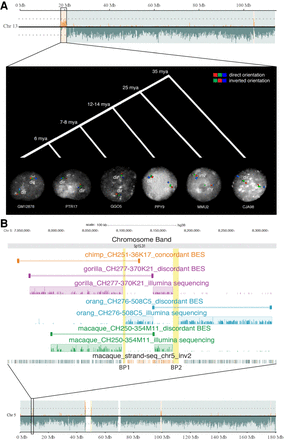
Evolutionary history of two inversions. (A) Strand-seq view of Chromosome 13 shows the switch in orientation of a 2-Mbp region, suggesting the presence of an inversion (Chr13_inv1). The region was tested using FISH in interphase nuclei in multiple primate species and was inverted just in macaque, whereas all the other primates are in direct orientation similar to human. (HSA) Homo sapiens; (PTR) Pan troglodytes; (GGO) Gorilla gorilla; (PPY) Pongo pygmaeus; (MMU) Macaca mulatta; (CJA) Callithrix jacchus. (B) Strand-seq view of a 89-kbp inversion (Chr5_inv2) between BP1 and BP2 is shown. BES mapping and Illumina sequencing of primate clones show that the region is inverted in gorilla, orangutan, and macaque and is direct in chimpanzee.











