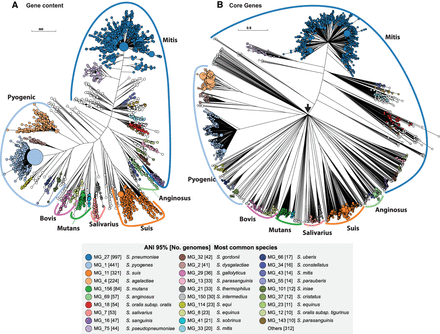
Phylogenies of 3052 Streptococcus genomes based on accessory gene content (A) and allelic variation in relaxed core genes (B). (A) A FastTree (Price et al. 2010) phylogeny based on binary information of the presence and absence of accessory genes. (B) A RapidNJ (Simonsen et al. 2011) phylogeny based on numbers of identical sequences (alleles) of 292 single copy, relaxed, core genes that are present in ≥95% of Streptococcus genomes. These trees are represented in GrapeTree (Zhou et al. 2018a). The sizes of the circles in A and B are proportional to the numbers of genomes they encompass and are color-coded by 29 common ANI95% clusters as shown in the inset. Many Streptococcus species have been assigned to one of six traditional taxonomic groups whose names are shown outside colored arcs. These trees define from the Suis group which contains Streptococcus suis. A black arrow in B shows the root of the tree, where multiple branches radiate directly outward owing to lack of resolution of cgMLST for such distant taxa. All ANI95% cluster information can be found in Supplemental Table S4. Interactive versions of the trees can be found at (A) https://achtman-lab.github.io/GrapeTree/MSTree_holder.html?tree=https://raw.githubusercontent.com/zheminzhou/PEPPA_data/master/Strep.content.json and (B) https://achtman-lab.github.io/GrapeTree/MSTree_holder.html?tree=https://raw.githubusercontent.com/zheminzhou/PEPPAN_data/master/Strep.CGAV.json.











