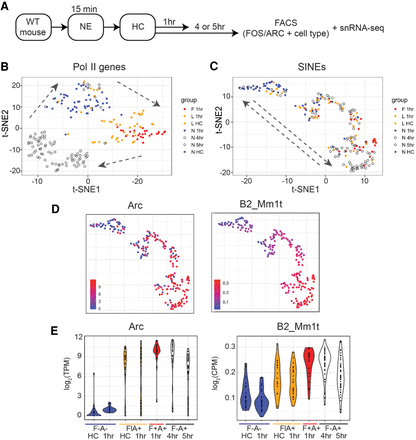
SINEs show IEG-like dynamics in DG neurons. (A) Study design from Jaeger et al. (2018), in which mice were raised in the HC, placed in a NE for 15 min, and then returned to the HC for 1, 4, or 5 h. The 4-h and 5-h nuclei were then sorted on FOS and ARC, as well as cell-type markers, and prepared for snRNA-seq. (B) T-SNE of DG nuclei from HC at the 1-, 4-, and 5-h time points using the standard RefSeq library of nonrepeat protein-coding and noncoding genes. Arrows denote the progression from an inactive state (blue; FOS−) to an early gene response (orange and red; FOS+) and then to a late gene response (open circles; FOS−ARC+). Orange, red = FOS+ nuclei with low or high levels of FOS protein stain during sorting, respectively. (C) T-SNE of the same DG nuclei in B, but based on the expression of bona fide SINEs. Arrows indicate the progression from an inactive state (blue; FOS−), through FOS+ low nuclei (orange), to FOS+ high (red), which coincides with late DG nuclei (FOS−ARC+). (D) The same t-SNE as in C, colored by Arc and B2_Mm1t expression. (E) Violin plots of Arc and B2_Mm1t expression by sorting group. (F) Fos, (A) Arc, (Fl) FOS low, (F+) FOS high.











