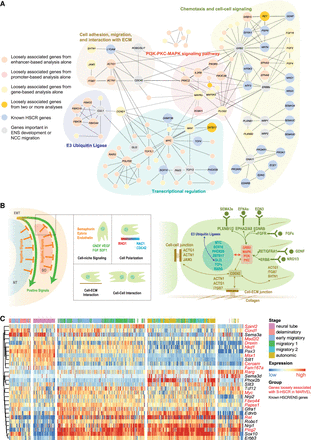
Functional landscape of S-HSCR-associated genes. (A) Functional interactions among genes loosely associated with S-HSCR, known HSCR genes, and genes important in ENS functions or NC migration. Each node corresponds to a gene and each edge corresponds to a functional interaction cataloged in Reactome. Genes of particular interest are shown in bigger nodes, labeled with their names. (B) Schematic illustration of some biological processes and genes involved in NC migration (Szabó and Mayor 2018). Colors of gene names follow their functional categories in panel A. (NT) Neural tube, (SO) somites. (C) Spatiotemporal expression profiles of mouse trunk NCs. Each row corresponds to the stage-specific expression pattern of a mouse homolog of a human gene shown in panel A. Genes identified by MARVEL as loosely associated with S-HSCR are shown in red. Each column corresponds to a single cell with the predicted stage label taken from the original publication (Soldatov et al. 2019). Both the rows and the columns were clustered using hierarchical clustering with Pearson's correlation as the similarity measure, and the columns are divided into five partitions according to the clustering results.











