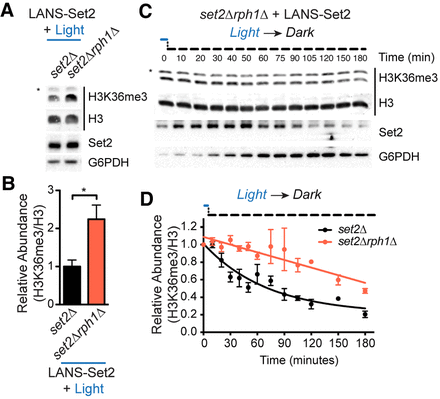
H3K36me3/2 removal is largely mediated by Rph1. (A) Representative western blot analysis of whole-cell lysates prepared from log-phase cultures of the indicated strains transformed with LANS-Set2 and grown continuously in the light. Asterisks indicate nonspecific bands. (B) Quantification of H3K36me3 from triplicate immunoblots shown in A and Supplemental Figure S5A. Data represent mean values ± SD (n = 3). (*) P < 0.05. (C) Representative western blot analysis of whole-cell lysates probing loss of H3K36me3 over time using LANS-Set2 in set2Δrph1Δ after the transition of log-phase cultures from light to dark (for replicates, see Supplemental Fig. S5E). Asterisks indicate nonspecific bands. (D) Quantification of H3K36me3 as a function of time from triplicate immunoblots shown in Figure 2G and Supplemental Figure S2B (set2Δ) and Figure 5C and Supplemental Figure S5E (set2Δrph1Δ). n = 3 and data represent mean ± SEM.











