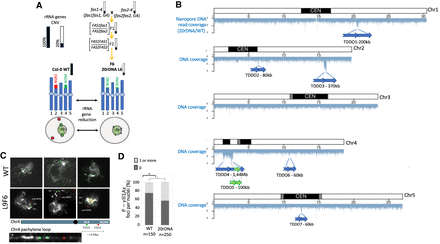
Genomic instability in the 20rDNA line L6F6. (A) Schematic representation of obtaining the 20rDNA L6F6 and its relative content in rRNA gene copies. NOR2 and NOR4 rRNA gene copies are both affected by the reduction, and according to the DNA-FISH experiment, it changes their nuclear distribution in the nucleus compared to wild-type Col-0 (WT), because all rRNA gene copies associate with the nucleolus (Pavlištová et al. 2016). (B) Distribution of the reads obtained by nanopore sequencing along all chromosomes in the 20rDNA L6F6 line compared to wild-type Col-0. The blue arrows represent the localization and the orientation of the TDDO identified in L6F6, except for TDDO5, which is represented by a green arrow. (C) DNA-FISH analyses of two loci present on the short arm of Chromosome 4 (kr4s), distant by 1.6 Mb: one present on the TDDO4 (BAC T5H22; green) and one located outside the TDDO4 (BAC F5I10; red). Three Pachytene chromosomes from WT Col-0 and in 20rDNA L6F6 are shown. A zoom of the unlooped kr4s from the middle panel of the 20rDNA line6 is presented at the bottom of the panel. (D) Histogram showing the percentage of nuclei displaying at least one P-γ-H2Ax foci in WT Col-0 versus L6F6.











