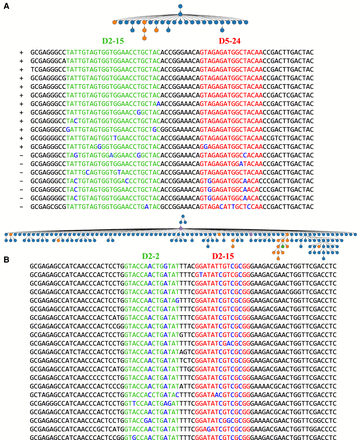
Clonal trees derived from V(DD)J recombinations in the INTESTINAL data set. Two large clonal trees derived from tandem fusions of D2-15 with D5-24 (A) and D2-2 with D2-15 (B) genes in the INTESTINAL data set. Genes D2-15 (AGGATATTGTAGTGGTGGTAGCTGCTACTCC), D5-24 (GTAGAGATGGCTACAATTAC), and D2-2*01 (AGGATATTGTAGTAGTACCAGCTGCTATGCC) have length 31, 20, and 31 nt, respectively (substrings occurring in CDR3s are underlined). A clonal tree for each clonal lineage was constructed using the IgEvolution tool (Safonova and Pevzner 2019b) applied to all reads from these lineages. These clonal lineages originated from a V(DD)J recombination that resulted in long CDR3s of length 72 and 78 nt, respectively. For the tree in B, we showed 20 out of 131 CDR3s. Blue, orange, and green vertices represent sequences of IgA memory, IgA plasma, and IgM plasma B cells, respectively. Violet vertices represent sequences found in both plasma and memory B cells. Alignments of CDR3s corresponding to the lineages are shown below the trees: green/red substrings correspond to fragments of D2-15/D5-24 genes in A and D2-2/D2-15 in B, respectively. Somatic hypermutations in green and red substrings are shown in blue. Plus and minus signs before each sequence in the alignment of the CDR3s in A indicates whether IgScout identified it as a tandem CDR3 or not (with the default k-mer-size parameter). CDR3s in B were semimanually annotated, as IgScout (with the default k-mer-size parameter) failed to identify them as tandem.











