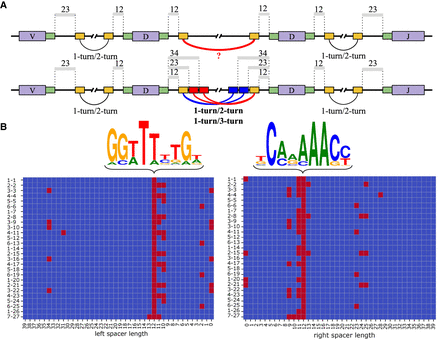
Cryptic nonamers explain V(DD)J recombination via the 1-turn/2-turn and 1-turn/3-turn mechanisms. (A) Canonical heptamers and nonamers in RSSs are shown by green and yellow rectangles, respectively. The 12/23 rule (1-turn/2-turn) explains the V-D and D-J recombination but fails to explain the D-D recombination using canonical nonamers (upper row). Cryptic nonamers (shown as red and blue rectangles) enable both the canonical 12/23 rule and the alternative 12/34 mechanism (1-turn/3-turn) and explain the V(DD)J recombination (lower row). (B) The left and right figures correspond to nonamers in the left and right RSSs. Sequence logos for canonical nonamers with 12-spacers for the human IGHD genes. Cryptic nonamers (with spacers shorter than 40 nt) in the RSSs of all 27 human D genes. D genes are shown on the left and are ordered according to the order in the IGHD locus. Canonical and cryptic nonamers (with likelihoods exceeding minLikelihood) are shown as red cells.











