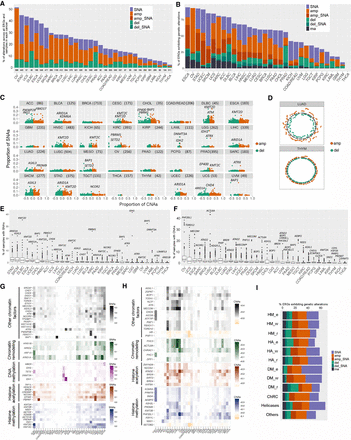
Pan-cancer analysis of genetic alterations across ERG categories and classes. (A,B) The percentage of samples with genetic deregulation in ERGs (A) and the percentage of ERGs showing different types of genetic deregulation (B), by cancer type. ERGs are considered altered if at least 1% of samples harbor these genetic aberrations. (C) Proportion of samples with SNAs versus that with deletions (−1, −2) or amplifications (+1, +2) in ERGs for each cancer type. Each gene is represented by two dots (red and green) depicting amplified and deleted CNAs, respectively. (D) Circos plots showing the relative amount of deregulation in CNAs by chromosomal distribution in two representative cancer types (LUAD and THYM, characterized by high and low CNA burden, respectively). The level of CNAs for each ERG was calculated as the proportion of samples considering all types of CNAs (amplification = +1, +2 and deletion = −1, −2) in ERGs in each cancer type. (E,F) Box plots showing the percentage of samples with SNAs (E) and deep CNAs (F) by gene and by cancer type. The most deregulated ERGs are highlighted for each cancer type. (G,H) Heatmaps representing the top genetically deregulated genes showing SNAs (G) and CNAs (H) in at least 10% and 15%, respectively, of the samples for any cancer type. Only samples with deep CNAs were included. ERGs are grouped into functional categories as indicated. (I) The percentages of ERGs that show genetic alteration among all cancer types by functional groups. Genetic alterations: (SNA) single-nucleotide alteration, (amp) deep copy number amplification, (amp_SNA) deep amplification co-occurring with SNA, (del) deep copy number deletion, (del_SNA) deep deletion co-occurring with SNA, and (ma) multiple alterations. In cases in which both types of CNAs (amplification and deletion) of one gene were present in the samples, we reported in B and H the alteration that was at least twice as prevalent as the other; otherwise, the alteration was reported under the multiple alteration category.











