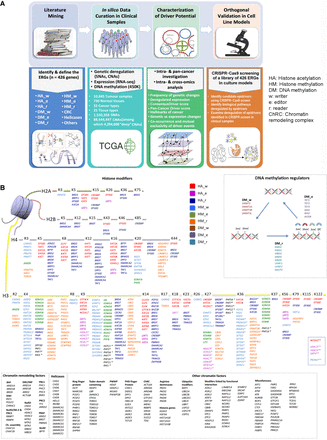
Study design. (A) A four-stage approach to identify and characterize ERGs with cancer driver potential. (B) The compendium of ERGs curated and analyzed, comprising 426 genes classified into histone modifiers, DNA methylation regulators, chromatin remodeling factors (ChRC), helicases, and other chromatin modifiers (some of which were further divided into subgroups based on function or their presence in molecular complexes). Histone acetylation, histone methylation, and DNA methylation modifiers are further stratified each into “writers” (w), “editors” (e), and “readers” (r). (*) The histone modifying genes whose functions are not well characterized and which were, therefore, assigned based on ENCODE ChIP sequencing data; (**) the histone modifying genes without assignment of residues in the histone tails.











