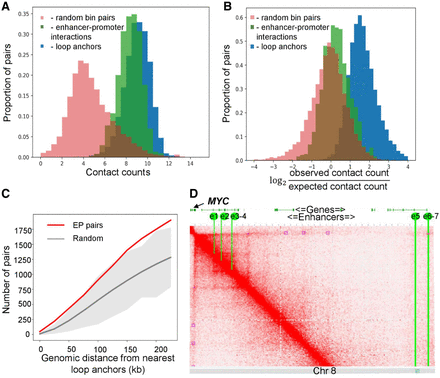
Hi-C loops do not provide complete information about interacting EP pairs. (A,B) Distribution of row (A) and distance-normalized (B) contact frequencies for interacting EP pairs and loop anchors in human monocytes. (C) Number of interacting EP pairs overlapping loops in monocyte data. Red line represents the number of EP pairs overlapping any Hi-C loop anchors or located within a distance not more than x kb of them, shown as a function of x. Gray line and gray area represent average plus three standard deviations of 100 randomized controls. (D) Chromatin interactions within the region on human Chromosome 8 containing seven experimentally validated MYC enhancers (yellow lines, e1–e7) and Hi-C loops (purple squares). Although both enhancers and loops were identified in the same cell type (K562 cells), they show little overlap.











