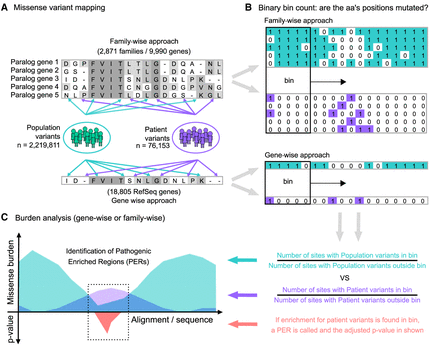
Study workflow and the PER viewer. (A) Starting from protein alignments of paralogous genes (gene-family approach) or all genes (gene-wise approach), missense variants from gnomAD (population; green) and ClinVar/HGMD (patient; purple) were mapped independently to the corresponding amino acid positions. (B) The mapping follows a binary notation. For sites with at least one missense variant reported, a “1” state was assigned. Alternatively, if no mutation was found, a “0” state was annotated instead. Amino acid sliding window (bin) counting over the alignment/sequence was used to calculate the corresponding missense burden. (C) The ratio between the number of sites with missense variants inside and outside the bin defines the burden area (population burden = green; patient burden = purple). Statistical comparison between the population and patient variant burden across aligned sequences allowed the identification of significant pathogenic variant enriched regions (PERs; red area).











