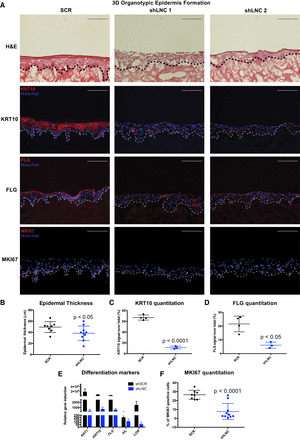
PRANCR is required for proliferation and differentiation in stratified epidermis. (A) Hematoxylin and eosin staining (top); immunofluorescence of the differentiation proteins KRT10 and FLG (middle), and immunofluorescence of proliferation marker MKI67 (bottom) in control and PRANCR-depleted (shLNC) epidermal tissue. Nuclei are stained in blue (Hoechst 33342). Scale bars, 100 µm. (B) Quantitation of epidermal thickness. Each dot represents the average of three measurements per image at fixed positions. Error bars, mean with SD; n = 8 in control and n = 10 in PRANCR knockdown. Differences evaluated using Student's t-test. (C,D) KRT10 and FLG quantitation as a percentage of the total fluorescence signal. Dots represent the average intensities measured from different images taken for each tissue. Error bars, mean with SD; n = 4 tissues in control and n = 3 tissues in PRANCR knockdown. (E) RNA expression in PRANCR-depleted epidermis versus control. Bars, mean with SEM; n = 4. (F) MKI67 quantitation as a percentage of total cells. Error bars, mean with SD; n = 8 in control and n = 10 in PRANCR knockdown. Differences evaluated using Student t-test. (SCR) Scrambled short hairpin; (shLNC1/2) short hairpin RNA 1 or 2 targeting PRANCR.











