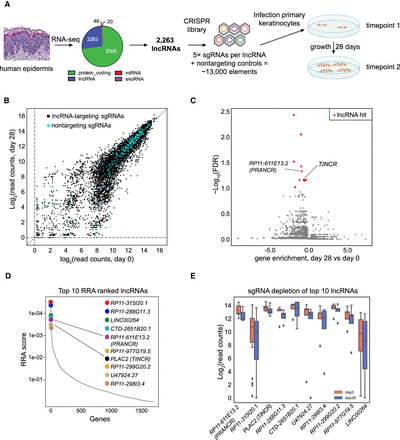
Discovery of lncRNAs controlling proliferation of epidermal progenitors. (A) Schematic of CRISPRi library design strategy and the CRISPRi screen. RNA sequencing identified 2263 lncRNAs with RPKM > 1. Five sgRNAs were designed for each target lncRNA transcript, together with 250 nontargeting controls, to form the CRISPRi library. (B) Scatter plots of sgRNA abundance at day 0 and day 28 of the screen. Nontargeting sgRNAs are shown in blue. (C) FDR values of each lncRNA candidate, as calculated by MAGeCK. A discovery threshold for positive hits (red) was defined by lncRNAs with similar or stronger FDR values as a known positive control, TINCR (previously known as PLAC2) (FDR threshold = <0.1). Gene enrichment represents average log-scale enrichment of sgRNAs changing concordantly with the lncRNA selection. (D) The robust ranking aggregation (RRA) scores of top lncRNA screen hits. (E) Normalized read counts of sgRNAs of the top 10 ranked lncRNA hits comparing postscreen (day 28) versus prescreen (day 0) abundance. Center lines represent median values; box limits represent the interquartile range; whiskers each extend 1.5 times the interquartile range; and dots represent outliers.











