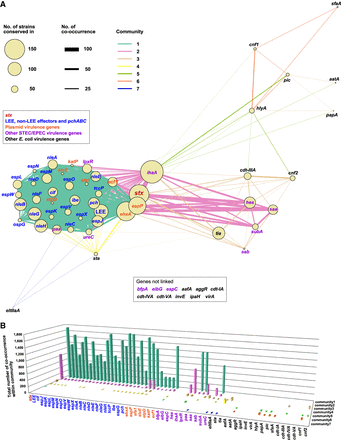
Co-occurrence network analysis of virulence genes. (A) Co-occurrence network analysis of virulence genes in bovine and human commensal isolates. Gene names are colored based on the virulence gene classification indicated in the box. The node sizes represent the number of strains in which each gene was conserved. Seven communities identified are indicated by differently colored edges (connections). Edge widths represent the number of co-occurrences. Only one co-occurrence between genes was excluded from the analysis. (B) Community members and their total numbers of co-occurrence within each community.











