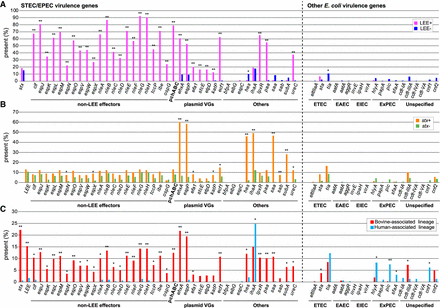
Conservation of STEC/EPEC-related and other E. coli virulence genes in bovine and human commensal isolates. Conservation of the virulence genes of STEC/EPEC and those of other E. coli pathotypes in LEE-positive (n = 104) and LEE-negative (n = 833) strains (A), in stx-positive (n = 146) and stx-negative (n = 791) strains (B), and in the bovine-associated (n = 647) and human-associated (n = 290) lineages (C) are summarized. Statistical analyses were performed using the two-sided Fisher's exact test. Asterisks indicate that the differences were significant after the Bonferroni correction for multiple comparisons: (*) P < 0.05; (**) P < 1 × 10−6.











