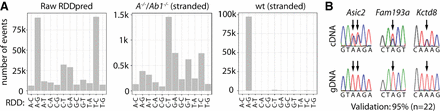
Nascent-seq identifies almost 100,000 mouse editing sites. (A) Nascent-RNA was prepared from brains of wild-type mice and subjected to Illumina sequencing (n = 3). After mapping to the mouse genome (mm10), potential RNA-DNA differences (RDDs) were determined using RDDpred (Raw RDDpred). To improve the identification of true RDDs, we removed all RDDs that either could not be mapped unambiguously to one strand, only occurred in one replica, or were also detected in an editing-deficient mouse line (Adar−/−, Adarb1−/−; A−/−, Ab1−/−). Thereby, we enriched for A-to-G RDDs indicative of A-to-I editing (wt stranded). (B) For validation, we used Sanger sequencing. Twenty-two editing sites were tested, and representative cDNA plus corresponding gDNA sequencing traces are shown (all traces are shown in Supplemental Fig. S2).











