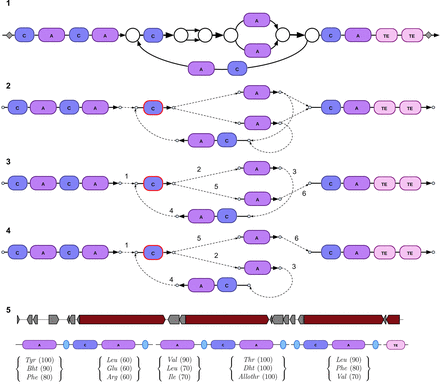
The BGC subgraph and the scaffolding graph for the supragingival plaque metagenome (SRS013723) in the HMP data set. (1,2) The BGC subgraph and the scaffolding graph. (3,4) Two rural postman routes in the scaffolding graph. The duplicated C-domain is highlighted with red border and is traversed twice in the rural postman routes. The numbers labeling the dashed edges indicate their order in the resulting tour. (5) Since biosyntheticSPAdes and antiSMASH use different thresholds and filtering options, antiSMASH identified only five (rather than six) A-domains in the NRP BGC predicted by biosyntheticSPAdes. The three most likely amino acids for each A-domain are shown along with their NRPSpredictor2 (Röttig et al. 2011) scores for the first of two rural postman routes.











