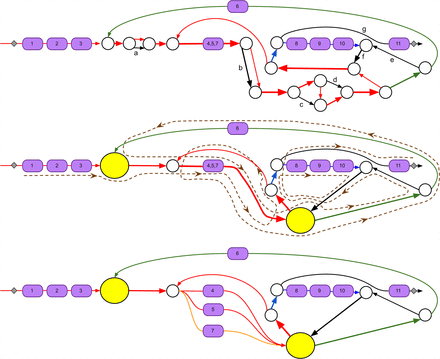
Subgraph of the assembly graph of S. coelicolor corresponding to the CALC NRP BGC. (Top) Edges of the assembly graph traversed by the CALC BGC. Nodes of the assembly graph are shown as white circles. After applying exSPAnder, the CALC BGC remains scattered over 10 scaffolds. Three of them are shown as red, blue, and green paths through the assembly graph; the remaining seven consist of a single edge each (shown in black and marked with letters a through g). The positions of eleven A-domains (with their indices) along the CALC BGC are shown by violet boxes. Edges with low and high coverage by reads are shown as thin and thick edges, respectively. The edge harboring three A-domains 4, 5, and 7 has approximately triple coverage by reads as compared to other domain-harboring edges. The 11 A-domains in CALC are split over three NRP synthetases with 6, 3, and 2 A-domains, respectively. (Middle) A simplified representation of the graph with all short edges (shorter than 300 bp) contracted into single vertices. The two contracted subgraphs of the assembly graph (formed by short edges) are represented by yellow vertices. The brown dashed path illustrates how the CALC NRP synthetase traverses the contracted assembly graph. (Bottom) The bubble restoration procedure described below transforms the collapsed edge harboring three A-domains (A-domains 4, 5, and 7) into three edges, each of them harboring a single A-domain. Applying exSPAnder to the modified assembly graph results in seven scaffolds that differ from scaffolds before bubble restoration (shown as red, blue, green, and orange paths as well as three black edges). Gray squares show the starting and ending positions of the CALC BGC.











