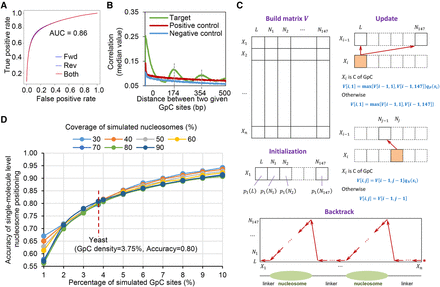
5mC detection and nucleosome occupancy detection by MeSMLR-seq data. (A) ROC curve of 5mC detection on GpC sites. The molecules that were aligned to forward (fwd) and reverse (rev) genomic strands were analyzed separately. (B) Correlation coefficients between methylation scores of mutually paired GpC sites from the same molecules with respect to their corresponding distances. (C) Dynamic programming algorithm for nucleosome occupancy detection (NP-SMLR). A matrix regarding the nucleotide sequence (row) and nucleosomal statuses (column) is made, followed by initialization, iterative update for entries, and backtrack search for optimal path (see Methods for details). (D) Accuracy of nucleosome occupancy detection under different nucleosome coverage and GpC frequencies.











