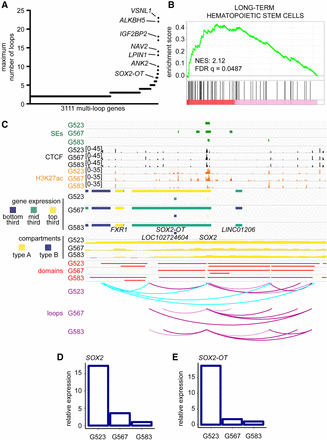
Interplay of 3D genome organization and chromatin features in transcriptional control of stemness genes in GBM. (A) Genes ranked by the number of loops they contact. Only genes with at least two loops are displayed. Loops called by HiCCUPS as 5-kb–100-kb resolution merged loops throughout this figure. (B) Gene set enrichment analysis based on gene ranking in A. (C) Integration of Genome Browser tracks for ROSE superenhancer calls, CTCF ChIP-seq, H3K27ac ChIP-seq, RNA-seq, compartments (50-kb), domains (10-kb), and loops (union of 5-, 10-, and 25-kb calls) determined by Hi-C at the SOX2 locus. Cyan arc tracks indicate a hub of culture-specific loops in G523. (D,E) Relative expression of SOX2 and SOX2-OT as determined by RNA-seq.











