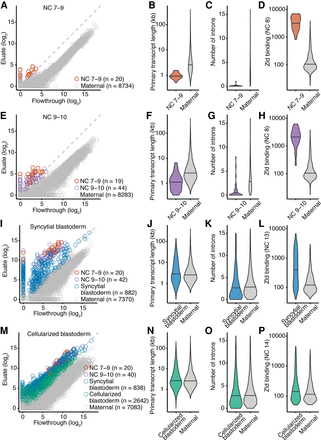
Gradual, continuous, and widespread activation of the zygotic genome. (A) Identification of 20 genes transcribed before pole-cell formation. Plotted for each gene is the level of mRNA in the eluate as a function of its level in the flowthrough. Genes passing the threshold for annotation as zygotically transcribed are indicated (red), as are those with detected transcripts that did not pass the threshold (gray). (B) Comparison of primary-transcript lengths of genes transcribed during NC 7–9 (red, n = 20) with those of genes with maternal transcripts (gray, n = 4873). Genes with maternal transcripts are those expressed in stage 14 oocytes (Kronja et al. 2014) and not annotated as zygotically transcribed in our study. (C) Comparison of intron numbers for genes transcribed during NC 7–9 (red, n = 20) with those for genes with maternal transcripts (gray, n = 4873). Maternal genes with intron numbers exceeding that of the 99th percentile were excluded. (D) Analysis of Zelda ChIP-seq signal, comparing the distribution of signals for genes transcribed during NC 7–9 (red) with that for genes with maternal transcripts (gray). Zelda ChIP-seq data were from embryos in NC 8 (Harrison et al. 2011). Of the genes transcribed during NC 7–9, all 20 had Zelda ChIP-seq signal, and of the set of 4873 genes with maternal transcripts, 494 had Zelda ChIP-seq signal. (E) Identification of 44 additional genes transcribed at NC 9–10. Points for genes identified as zygotically transcribed in two biological replicates are colored, indicating in purple those not identified in the previous stage. Otherwise, this panel is as in A. (F–H) As in B–D but comparing results of the additional genes transcribed at NC 9–10 (purple) with those of genes with maternal transcripts. Of the 44 genes newly transcribed during NC 9–10, 40 had Zelda ChIP-seq signal. (I) Identification of 882 additional genes transcribed in NC 11–13 (blue). Otherwise, this panel is as in E. (J–L) As in B–D but examining the additional genes transcribed in the syncytial blastoderm (blue). Zelda ChIP-seq data were from embryos at NC 13. Of the 882 genes newly transcribed in the syncytial blastoderm, 336 had Zelda ChIP-seq signal at NC 13, and of the set of 4873 genes with maternal transcripts, 567 had Zelda ChIP-seq signal. (M) Identification of 2642 additional genes transcribed in NC 14 (green). Otherwise, this panel is as in I. (N–P) As in B–D examining the additional genes transcribed in the cellularized blastoderm (green). Zelda ChIP-seq data were from embryos at NC 14. Of the 2642 genes newly transcribed in the cellularized blastoderm, 511 had Zelda ChIP-seq signal, and of the set of 4873 genes with maternal transcripts, 679 had Zelda ChIP-seq signal. All pairs of violin plots were scaled to have the same area. Horizontal line denotes median.











