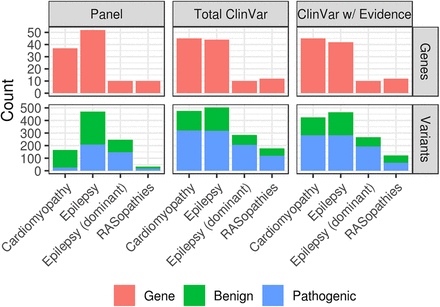
Study data sets. Missense variant and gene counts are shown by disease panel and ClinVar variant set. We only used ClinVar variants from panel genes and considered either any ClinVar variant (Total ClinVar), or ClinVar variants that have been reviewed (ClinVar w/Evidence). ClinVar variants were restricted to those with no conflicting pathogenicity assignments, and any genomic position from the panel data was removed from the ClinVar variant sets.











