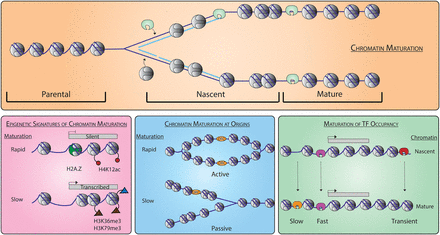
Genome-wide chromatin maturation behind the DNA replication fork. Parental chromatin is re-established behind the replication fork (top). Locus-specific differences in chromatin maturation possess different epigenetic signatures (bottom left). Chromatin maturation at start sites of DNA replication is different for active and passive origins (bottom middle). Transcription factors (TFs) associate with different kinetics behind the DNA replication fork including sites of transient occupancy (bottom right).











