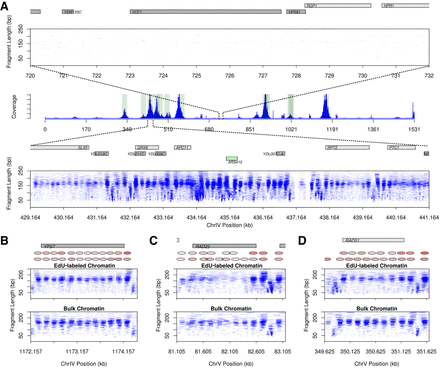
Enrichment of EdU-labeled chromatin at sequences proximal to early origins. (A) Sequencing coverage across Chromosome IV. NCOP at an early replicating origin shows a defined chromatin architecture (bottom) compared with a nonreplicated region (top). (Middle) Enrichment of EdU-labeled sequences along Chromosome IV. Shaded green background highlights early origins of replication (Belsky et al. 2015). (Bottom) The green box represents an origin of replication. In the case of the region depleted of nucleosomes at ∼437 kb, there is a transposable element (YDLWtau1 [Ty4 LTR]) that overlaps this position. (B–D) Chromatin profiles at genes proximal to an early origin that showed unaltered (B) and altered (C,D) chromatin structure. Nucleosome position and occupancy are depicted with red shaded ovals. Gray boxes represent genes on the positive (light gray) and negative (dark gray) strands. (D) RAD51 shows recruitment of a transcription factor and downstream nucleosome shift in the EdU pull-down experiment compared with total chromatin.











