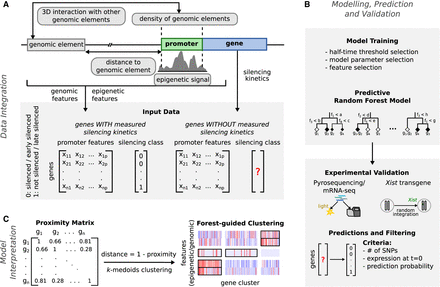
Schematic overview of our modeling approach. (A) Epigenetic and genomic input data for the model are collected, and feature matrices are computed for all X-linked genes with estimated half-times (labeled) and without estimated half-times (unlabeled). (B) After model training, the XCI/escape model is then used to predict the silencing class of all unlabeled X-linked genes given the same set of input features. The predictions are validated by comparing them to measured half-times from undifferentiated mRNA-seq data, with pyrosequencing experiments (few selected genes) and with measured silencing dynamics of genes in six transgenic mESCs clones. (C) A forest-guided clustering approach was developed for model interpretation. A proximity matrix between genes is computed from the trained model and converted into a distance matrix. Clusters of genes and their most significant associated features are displayed as a heatmap.











