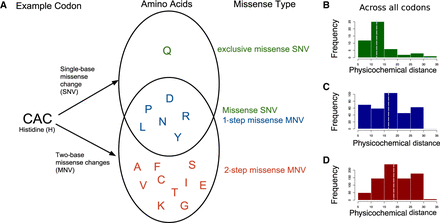
Classification of intra-codon MNV missense mutations. (A) Example of how one-step missense MNVs and two-step missense MNVs are classified using a single codon “CAC.” Venn diagram shows amino acids that can be created with either a single-base change or a two-base change in the codon “CAC.” (B–D) Across all codons, the distribution of physicochemical distances for the amino acid changes caused by different types of missense variants: (B) exclusive SNV missense; (C) one-step MNV missense; and (D) two-step MNV missense. Dashed line indicates the median of the distribution.











