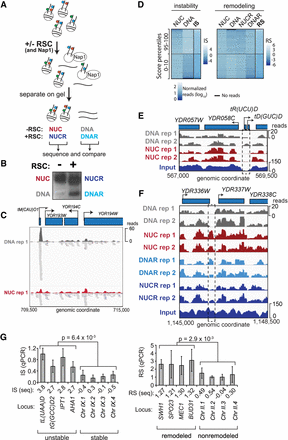
Chromatin remodeling and instability studies performed on native chromatin. (A) Schematic of the nucleosome disassembly assay. (B) Nucleosomes incubated +/− RSC were separated by native agarose gel electrophoresis. The four indicated bands were excised and the DNA extracted and sequenced. (C) Representative example of highly unstable nucleosome at tM(CAU)O1 (Chromosome XV), enriched in the ‘DNA’ band after incubation but also detected in ‘NUC.’ (D) Heat maps of read densities for nucleosomes with different IS and RS, respectively, comparing distinct percentiles. Left boxes, distribution of read densities in bands NUC and DNA for corresponding IS distributions. Right box, equivalent distribution for bands used to calculate RS; 10,000 randomly sampled windows in the 10th and 95th percentiles are shown. (E) Region encompassing a representative unstable nucleosome (dashed box; Chromosome IV [IS = 1.0]). (F) As E, but for a strongly remodeled nucleosome (Chromosome IV [RS = 0.78]). (G) qPCR validation, using primers for the indicated regions. Left, plot of instability index for qPCR (IS-qPCR). Four unstable nucleosomes followed by four stable nucleosomes. The P-value was calculated using the Wilcoxon t-test; error bars show standard deviations from four biological replicates. Right, as left, but for remodeling. Three biological replicates were performed.











