Convergent recombination cessation between mating-type genes and centromeres in selfing anther-smut fungi
- Fantin Carpentier1,4,
- Ricardo C. Rodríguez de la Vega1,4,
- Sara Branco1,6,
- Alodie Snirc1,
- Marco A. Coelho2,7,
- Michael E. Hood3,5 and
- Tatiana Giraud1,5
- 1Ecologie Systématique Evolution, Bâtiment 360, Univ. Paris-Sud, AgroParisTech, CNRS, Université Paris-Saclay, 91400 Orsay, France;
- 2UCIBIO-REQUIMTE, Departamento de Ciências da Vida, Faculdade de Ciências e Tecnologia, Universidade NOVA de Lisboa, 2829-516 Caparica, Portugal;
- 3Department of Biology, Amherst College, Amherst, Massachusetts 01002, USA
-
↵4 These authors contributed equally to this work.
Abstract
The degree of selfing has major impacts on adaptability and is often controlled by molecular mechanisms determining mating compatibility. Changes in compatibility systems are therefore important evolutionary events, but their underlying genomic mechanisms are often poorly understood. Fungi display frequent shifts in compatibility systems, and their small genomes facilitate elucidation of the mechanisms involved. In particular, linkage between the pre- and postmating compatibility loci has evolved repeatedly, increasing the odds of gamete compatibility under selfing. Here, we studied the mating-type chromosomes of two anther-smut fungi with unlinked mating-type loci despite a self-fertilization mating system. Segregation analyses and comparisons of high-quality genome assemblies revealed that these two species displayed linkage between mating-type loci and their respective centromeres. This arrangement renders the same improved odds of gamete compatibility as direct linkage of the two mating-type loci under the automictic mating (intratetrad selfing) of anther-smut fungi. Recombination cessation was found associated with a large inversion in only one of the four linkage events. The lack of trans-specific polymorphism at genes located in nonrecombining regions and linkage date estimates indicated that the events of recombination cessation occurred independently in the two sister species. Our study shows that natural selection can repeatedly lead to similar genomic patterns and phenotypes, and that different evolutionary paths can lead to distinct yet equally beneficial responses to selection. Our study further highlights that automixis and gene linkage to centromeres have important genetic and evolutionary consequences, while being poorly recognized despite being present in a broad range of taxa.
Mating systems reflect the degree of selfing/outcrossing in natural populations and impact gene flow, the accumulation of deleterious alleles, and adaptability (Lande and Schemske 1985; Charlesworth and Charlesworth 1987; Charlesworth et al. 1990; Charlesworth 2002; Igic et al. 2008; Hereford 2010; Lande 2015). Outcrossing can promote gene flow and therefore the rapid spread of beneficial alleles as well as the purge of deleterious alleles, whereas selfing is often associated with reproductive assurance and can help maintain favorable combinations of alleles at different loci. There is a wide diversity of mating systems in nature that strongly impacts the evolution of organisms. Automixis with mating among products of a given meiosis that separated in the first meiosis division (“fusion of nonsister second division products”) (Lewis and John 1963), for example, is a little-known form of self-fertilization (Mogie 1986); such automixis is often called central fusion in animals, the term “fusion” referring to the union of gametes and the term “central” referring to the placement of the fusing gametes in an ordered tetrad (Suomalainen 1950; Goudie and Oldroyd 2014). This kind of automixis maintains heterozygosity at all loci for which there has been no recombination with the centromere (Hood and Antonovics 2000, 2004; Zakharov 2005; Lenormand et al. 2016; Engelstädter 2017). This effect can extend over large portions of the genome when there are low levels of crossing-over (Hood and Antonovics 2000, 2004). Automixis with central fusion can thus maintain long-term heterozygosity, which can lead to sheltering deleterious alleles or may be beneficial in cases of advantageous overdominance (i.e., heterozygote advantage) (Engelstädter 2017). Automixis and its genetic and evolutionary consequences are poorly studied despite being relatively frequent (Mogie 1986) across a variety of taxa such as in fungi (Hood and Antonovics 2000; Zakharov 2005; Menkis et al. 2008; Grognet et al. 2014), plants (Asker 1980; Walker 1985; Antonius and Nybom 1995; Cruden and Lloyd 1995; Schön et al. 2009), reptiles (Watts et al. 2006; Booth et al. 2011; Booth and Schuett 2015), fishes (Chapman et al. 2007; Dudgeon et al. 2017; Feldheim et al. 2017), birds (Schut et al. 2008), crustaceans (Nougué et al. 2015), nematodes (Van der Beek et al. 1998), and insects (Suomalainen et al. 1976; Normark 2003; Oldroyd et al. 2008).
Evolutionary transitions between mating systems are known to be relatively frequent (Goldberg et al. 2010; Goldberg and Igić 2012; Chantha et al. 2013; Nieuwenhuis et al. 2013; Hanschen et al. 2018). Changes in the genetic determination of gamete production or compatibility often underlie transitions in mating systems, such as the evolution of a self-incompatibility system. For example, in many species mating can only occur between males and females, which enforces outcrossing, and sexes are often determined by sex chromosomes (Beukeboom and Perrin 2014). In angiosperms, mating can also be restricted by a self-incompatibility locus, which promotes outcrossing in hermaphroditic species by preventing mating between genotypes carrying identical alleles (Vekemans et al. 2014). In most fungi, gamete compatibility is controlled at the haploid stage, and only cells carrying different alleles at the mating-type loci can successfully mate (Billiard et al. 2011, 2012).
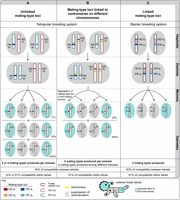
Fungi provide excellent eukaryotic models for studying the genomic changes involved in gamete compatibility transition, because they display highly diverse and labile mate-recognition systems (Billiard et al. 2011, 2012; Nieuwenhuis et al. 2013) as well as relatively small and compact genomes that allow for high-quality genome assemblies (Gladieux et al. 2014; Badouin et al. 2015; Faino et al. 2015; Sonnenberg et al. 2016; Branco et al. 2017, 2018; Sun et al. 2017a,b). In basidiomycete fungi (e.g., rusts, smuts, and mushrooms), mating type is most often controlled by two loci: (1) the PR locus, determining gamete fusion compatibility with a pheromone receptor and neighboring pheromone genes; and (2) the HD locus, determining compatibility for postmating development with two homeodomain genes (Coelho et al. 2017). To successfully mate and produce offspring, two gametes must carry different alleles at both loci. In most basidiomycetes, the PR and HD loci segregate independently (Raper 1966; Nieuwenhuis et al. 2013). Multiple independent events of linkage of the two mating-type loci have been documented in several fungal species (Bakkeren and Kronstad 1994; Nieuwenhuis et al. 2013; Branco et al. 2017; Sun et al. 2017b). Such control of gamete compatibility inherited as a single locus is advantageous under selfing because it increases the odds of gamete compatibility among the gametes of a given diploid individual (Fig. 1; Coelho et al. 2017).
Odds of compatibility among gametes of a diploid individual in basidiomycete fungi. Gametes are fully compatible only if they carry different alleles at both mating-type loci, the PR (including pheromone receptor and pheromone genes, with a1 and a2 alleles) and HD (including homeodomain genes, with b1 and b2 alleles) loci. (A) With PR and HD mating-type loci unlinked from each other and from the centromeres (shown here located in different chromosomes in blue and red), the percentage of compatibility of a given gamete among the other gametes produced by the same diploid individual is 25% across multiple meioses (a given gamete is compatible with one of every four gametes), and the percentage is 33% within tetrad (a given gamete is compatible with one of the other three gametes in the tetrad) or 67% (a given gamete is compatible with two of the three remaining gametes in the tetrad), depending on segregation of the mating-type alleles. The different types of gametes produced are tetratypes (TT), parental ditypes (PD), or nonparental ditypes (NPD), which depend on allele segregation and on whether a crossing-over occurred between one of the two loci and the centromere. (B) With PR and HD mating-type genes linked to the centromeres of different chromosomes (blue and red), the percentage of compatibility of a given gamete among the other gametes produced by the same diploid individual is 25% across multiple meioses but 67% within a tetrad (a given gamete is compatible with two of the three other gametes in the tetrad) due to the segregation of the variation occurring only at meiosis I for both mating-type loci. The different types of gametes produced are parental ditypes (PD) or nonparental ditypes (NPD), which depend on segregation. (C) With HD and PR loci fully linked to each other on the same chromosome, the percentage of compatibility of a given gamete among the other gametes produced by the same diploid individual is 50% across multiple meioses (a given gamete is compatible with one of every two gametes), and 67% within a single meiotic tetrad (a given gamete is compatible with two of the three other gametes in the tetrad). The light blue background shows the opposite cells of ordered tetrads, both in a Microbotryum linear tetrad representation (bottom) and in the different types of possible tetrads depicted depending on mating-type locus linkage.
The plant-castrating anther-smut fungi belonging to the highly selfing basidiomycete genus Microbotryum are particularly good systems for studying the genomic changes underlying shifts in gamete compatibility systems. Before the radiation of this genus, recombination suppression extended around each of the PR and HD loci (Branco et al. 2017). Several Microbotryum species underwent independent transitions to complete linkage between the mating-type loci through various chromosomal rearrangements that brought the HD and PR loci onto the same chromosome (Branco et al. 2017, 2018). In some of these species, the cessation of recombination subsequently expanded far beyond the mating-type loci in several successive steps to include the majority of the mating-type chromosomes (Branco et al. 2017, 2018). Recombining pseudoautosomal regions (PARs) remained at both edges of the mating-type chromosomes in many lineages (Branco et al. 2018).
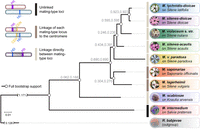
The majority of studied anther-smut fungi undergo selfing by automixis (Hood and Antonovics 2000; Giraud et al. 2008; Vercken et al. 2010; Gladieux et al. 2011; Bueker et al. 2016) and have linked mating-type loci (Branco et al. 2018). Here, we studied two closely related species, Microbotryum lagerheimii and Microbotryum saponariae, that have retained unlinked PR and HD mating-type loci located on different chromosomes (Fig. 1; Hood et al. 2015), despite selfing mating systems (Fortuna et al. 2016, 2018; Abbate et al. 2018). However, M. saponariae displays the PR and HD mating-type loci completely linked to the centromere of their respective chromosomes, which induces central fusion automixis and ensures the same odds of compatibility under selfing by automixis as would linkage between the mating-type loci (Fig. 1; Hood et al. 2015). Although the mating-type loci are also known to be located on different chromosomes in M. lagerheimii (Branco et al. 2017), it is unclear whether the HD and PR loci are linked to the centromeres. In case they are linked to centromeres and given that M. lagerheimii and M. saponariae are sister species in available phylogenies (Fig. 2), recombination cessation with the centromeres could potentially predate their speciation event. An alternative hypothesis would be independent linkage events, with convergence for complete centromere linkage occurring in the two species after their divergence. In this study, we used segregation analyses and high-quality genome assemblies to investigate (1) whether HD and PR loci are linked to the centromeres in M. lagerheimii, (2) whether linkage predates speciation between M. lagerheimii and M. saponariae or constitutes independent events, and (3) whether the PR and HD loci became linked to centromeres at similar dates in each species.
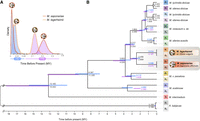
Phylogenies of anther-smut fungi and their mating-type loci linkage. Microbotryomycete phylogenetic tree based on 780 orthologous genes, including the studied Microbotryum species (shown in the anthers of their host plants) and the outgroup Rhodotorula babjevae. The empty circles indicate full bootstrap support. Tree internode certainty with no conflict bipartitions (the normalized frequency of the most frequent bipartition across gene genealogies relative to the summed frequencies of the two most frequent bipartitions) is given above the branches, indicating good support for the bipartitions. Black bars at right indicate unlinked mating-type loci, dark gray linkage of mating-type loci to centromeres, and light gray mating-type loci linkage.
Results
Linkage of mating-type loci to centromeres in M. saponariae and M. lagerheimii
To test whether recombination was suppressed between the mating-type loci and their respective centromere in M. lagerheimii, we analyzed PR and HD mating-type loci segregation within ordered linear tetrads using allele-specific PCR markers for each mating-type locus. When there is complete centromere linkage, alleles at both mating types always segregate at the first meiotic division, leading to the ordered linear Microbotryum tetrad with cells derived from opposite poles of meiosis I carrying alternate alleles at both the PR and HD loci (Fig. 1; Hood and Antonovics 2000; Hood et al. 2015). Conversely, when there is no centromere linkage of the mating-type loci, mating-type alleles segregating at the second meiotic division results in only half the tetrads carrying alternate alleles at both loci in the opposite cells of the ordered linear tetrad (Fig. 1). We found evidence supporting centromere linkage of mating-type loci in M. lagerheimii, with isolated meiotic products derived from opposite poles of meiosis I showing alternate alleles at both the PR and HD loci in all of the 78 meioses analyzed. Given the number of tetrads analyzed, the 95% confidence interval for the occurrence of recombination between at least one mating-type locus and its centromere was 0%–5%. This indicates that the M. lagerheimii PR and HD loci are completely or nearly completely linked to their respective centromere, as in its sister species M. saponariae (Hood et al. 2015).
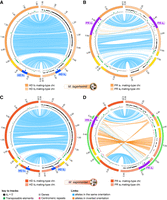
We compared the sequences of the mating-type chromosomes to investigate whether inversions could have contributed to linkage between mating-type loci and their centromeres. While the alternate HD mating-type chromosomes were collinear within both M. saponariae and M. lagerheimii (Fig. 3A,C), we observed a 51.4 kbp inversion between HD and the centromere in the M. saponariae lineage compared to the ancestral state, shared by Microbotryum intermedium and M. lagerheimii (Supplemental Figs. S2A, S3A). In both M. saponariae and M. lagerheimii, the HD locus was located close to the centromere (distant by 138 kbp in M. saponariae and by 162 kbp in M. lagerheimii) (Fig. 3A,C). In M. lagerheimii the alternate PR chromosomes (a1 and a2) also showed nearly complete collinearity (Fig. 3B), with only rearrangements around the PR locus, as typical in Microbotryum due to very old recombination suppression in this region (Branco et al. 2017). In contrast, M. saponariae displayed a large pericentric inversion distinguishing the alternate PR chromosomes. This inversion involved 593 kbp in the a1 and 701 kbp in the a2 PR mating-type chromosomes, representing 50% and 52% of a1 and a2 PR chromosome lengths (Fig. 3D). The two edges of the inversion were very close to the centromere and the PR locus (inversion boundaries were distant by 35 kbp from centromeres and by 0 kbp from the edge of the PR-proximal region with ancient recombination suppression) (Fig. 3D). The inversion appeared derived in the a2 M. saponariae PR chromosome, as the M. saponariae a1 PR chromosome was highly collinear to the M. lagerheimii a1 and a2 PR chromosomes (Supplemental Fig. S1). No further rearrangements were present within the large inversion beyond those located in the PR-proximal region (Fig. 3D). A small additional inversion was observed toward the PAR in the short arm of the M. saponariae a2 PR mating-type chromosome (green region in Fig. 3D, involving 11 kbp and 20 kbp on the a1 and a2 PR mating-type chromosomes). This inversion may correspond to an additional step extending further recombination cessation toward the PAR.
Intraspecific comparison of gene order between mating-type chromosomes. Comparison of gene order between HD and PR chromosome pairs in M. lagerheimii (A,B) and M. saponariae (C,D). The outer tracks represent contigs, staggered every 200 kb. The HD, PR, and pheromone genes are indicated by blue, dark purple, and small light purple circles, respectively. Blue and orange lines link alleles, the latter corresponding to inversions. The link width is proportional to the corresponding gene length. Yellow regions on the contig track indicate the centromeres (regions with low gene density, high TE density, and enriched in tandem-repeats marked in pink). The black marks along the right contigs track indicate genes that have no synonymous substitutions between a1 and a2 alleles within species (dS = 0). Green marks indicate transposable elements (TEs), and gray marks non-TE genes. The ancient regions of recombination suppression are indicated on the outer track in blue for the HD locus and in purple for the PR locus. (A) Comparison of the b1 (left, orange) and b2 (right, light orange) HD M. lagerheimii mating-type chromosomes. (B) Comparison of the a1 (left, orange) and a2 (right, light orange) PR M. lagerheimii mating-type chromosomes. (C) Comparison of the b1 (left, red) and b2 (right, light red) HD M. saponariae mating-type chromosomes. (D) Comparison of the a1 (left, red) and a2 (right, light red) PR M. saponariae mating-type chromosomes. The large green arrow indicates the large inversion between the two mating-type chromosomes encompassing the mating-type locus and the centromere. The green regions on the contig track of each mating-type chromosome indicate the small inversion that likely occurred after the large inversion linking the PR locus to the centromere, extending the region of suppressed recombination.
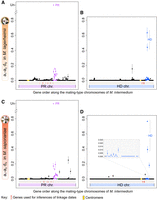
Synonymous divergence (dS) between alleles associated to the alternative mating types at the genes located between the PR-proximal region and the centromere in M. lagerheimii and M. saponariae provided further evidence for complete centromere linkage of the PR mating-type loci. For all genes linked to a mating-type locus, the same allele remains associated with the same mating-type, so that alleles associated to the alternate mating types accumulate independent mutations, showing increasing divergence (dS) with time since the complete recombination cessation. To recover the history of recombination cessation, we plotted allelic divergence (dS) along the ancestral gene order, as subsequent rearrangements may blur historical steps (Branco et al. 2017). We used the mating-type chromosomes of an outgroup species with independently segregating mating types (M. intermedium) as a proxy for the ancestral gene order; however, using the M. lagerheimii gene order gave similar conclusions given the few rearrangements observed (Supplemental Figs. S2, S3; Branco et al. 2017). We observed very high levels of synonymous divergence around the PR and HD mating-type loci both in M. lagerheimii and M. saponariae (purple and blue genomic regions, respectively) (Fig. 4). This result was expected given ancient recombination cessation proximal to each of the PR and HD mating-type loci (Branco et al. 2017). The nonzero dS values between the PR-proximal region and the centromere in M. saponariae and M. lagerheimii supported complete linkage to the centromere. In highly selfing organisms such as anther-smut fungi, homozygosity is high at almost all genes (i.e., dS = 0 between alleles on autosomes in a diploid individual) (Supplemental Fig. S4) except in regions linked to mating types (Branco et al. 2018). The lower dS values closer to the centromere rather than closer to the purple region, in both M. saponariae and M. lagerheimii (Fig. 4A,C), suggest stepwise extension of recombination cessation farther from the PR locus and to eventually reach the centromere. The synonymous divergence between the HD-proximal blue region and its centromere in both species was almost zero, although some genes exhibited nonzero dS value (Fig. 4B,D).
Per-gene synonymous divergence and respective standard error (dS ± SE) between alleles associated to the a1b1-a2b2 mating types along the mating-type chromosomes within diploid M. lagerheimii and M. saponariae individuals. Synonymous divergence is plotted against the genomic coordinates of the a1 b1 mating-type chromosomes of M. intermedium for all single-copy genes shared by the mating-type chromosomes, as a proxy for ancestral gene order. Divergence between the a1 and a2 pheromone receptor (PR) was too extensive (Devier et al. 2009) and could not be computed (noted as unalignable [UN]). The positions of the centromeres are indicated by yellow dots. Genes with dS > 0 between mating types around the PR and HD mating-type loci in M. lagerheimii are in purple and blue, respectively. The purple and blue regions correspond to the older PR and HD regions of suppressed recombination that evolved before and at the base of the radiation of the clade, respectively (Branco et al. 2017, 2018). Red arrows indicate the genes used for dating recombination cessation events. (A) Per-gene synonymous divergence between mating types in M. lagerheimii along the gene order of the a2 PR M. intermedium mating-type chromosome. (B) Per-gene synonymous divergence between mating types in M. lagerheimii along the gene order of the b2 HD M. intermedium mating-type chromosome. (C) Per-gene synonymous divergence between mating types in M. saponariae along the gene order of the a2 PR M. intermedium mating-type chromosome. (D) Per-gene synonymous divergence between mating types in M. saponariae along the gene order of the b2 HD M. intermedium mating-type chromosome.
We found increased transposable element content in the HD and PR chromosomes in M. saponariae and M. lagerheimii compared to their autosomes. There were differences within each species between a1 and a2 mating-type chromosomes (Supplemental Fig. S5). Such transposable element accumulation and differences between homologous chromosomes further supported complete recombination cessation.
Absence of trans-specific polymorphism between centromeres and the HD- and PR-proximal regions in M. saponariae and M. lagerheimii
We used genealogies of genes located between centromeres and the HD- and PR-proximal blue and purple regions, respectively, to assess whether linkage of mating-type loci to centromeres in M. saponariae and M. lagerheimii derived from a single event predating their speciation or from independent events in each lineage. If recombination cessation predates speciation, the alleles associated to the alternative mating types will cluster by mating type rather than by species (which is called trans-specific polymorphism), because the alleles will have been linked to mating types since before the speciation. In contrast, if linkage is more recent than speciation, alleles will cluster by species because recombination will have broken any allelic association with mating-type within species after speciation. None of the orthologous groups corresponding to genes located between the PR- or the HD-proximal purple and blue regions and their centromeres displayed trans-specific polymorphism shared by M. saponariae and M. lagerheimii. We could use nine genes in the HD mating-type chromosome (Supplemental Fig. S6A) and 10 genes in the PR mating-type chromosomes (Supplemental Fig. S6B), for which both alleles were available in all species with available genomes (the genes are indicated by red arrows on Fig. 4). These findings indicate independent events of complete mating-type-loci-centromere linkage in M. saponariae and M. lagerheimii.
We found further support for recombination suppression occurring after M. saponariae and M. lagerheimii speciation by dating the differentiation between alleles associated with a1 versus a2 mating types in gene genealogies. We computed a phylogenetic tree using the concatenated alignments of the nine and 10 genes with both alleles available in all genomes and located between the HD-proximal region and the centromere, or the PR-proximal region and the centromere, respectively. We calibrated the tree nodes using the speciation date between Microbotryum lychnidis-dioicae and Microbotryum silenes-dioicae, previously estimated at 420 ky (Gladieux et al. 2011). Although these estimates are not robust absolute dates, they are useful to obtain relative dates of speciation and chromosome evolution events. Recombination cessation between the PR-proximal purple region and the centromere was younger in M. saponariae and M. lagerheimii (95% confidence interval 171–302 and 80–158 ky, respectively) (Fig. 5A) than their speciation event (95% confidence interval 2997–4386 ky) (Fig. 5B). The date of recombination cessation between the HD-proximal blue region and the centromere was even younger in both species (95% confidence interval 0.1–18 and 3–26 ky, respectively) (Fig. 5A).
Linkage date estimates across Microbotryum. Linkage between mating-type loci (PR and HD) and centromeres, or between PR and HD loci, was inferred from dates of divergence between alleles associated to the a1 and a2 mating types at genes linked to the mating-type loci. In gene genealogies, nodes separating alleles associated to the a1 and a2 mating types within species correspond to the date of linkage to the mating-type loci (to the HD locus in blue and to the PR locus in purple). Genealogies of alleles associated to the alternative mating types (a1 and a2) were reconstructed based on a concatenated alignment of both alleles at nine genes ancestrally located between the centromere and the HD-proximal region (8525 aligned codons) and of 10 genes ancestrally located between the centromere and the PR-proximal region (10,200 aligned codons). The genes used for this analysis, with both alleles in all species, are indicated by red arrows in Figure 4. (A) Marginal posterior densities for the most recent common ancestor date (MRCA time) estimated with BEAST v2.4.0 based on multiple sequence alignments of genes located between the HD-proximal (blue) or the PR-proximal (light purple) region and the corresponding centromeres in the ancestral gene order. (B) Time-calibrated tree of a1 and a2 alleles with nodes drawn at the mean date (in million years [MY]) for the genes on the HD chromosome. Inferred divergence dates for the genes located between the HD-proximal or the PR-proximal regions and the corresponding centromeres are shown in blue or purple fonts, respectively, to the right of the nodes. Light blue and light purple bars correspond to 95% confidence intervals. Speciation dates as inferred from each data set are shown in black font on the right side of the nodes (bottom, PR set; top, HD set).
Discussion
Here, we document convergent evolution of increased odds of gamete compatibility under automixis by independent linkage events of mating-type loci and centromeres in two closely related fungal species. Such linkage represents further convergence in gamete compatibility patterns with other congeneric lineages, which were previously shown to have achieved similar gamete compatibility odds through multiple independent direct linkage events between PR and HD mating-type loci (Branco et al. 2018). Linkage of the two mating-type loci, one to each other or to their centromere, are equally beneficial under automixis in terms of gamete compatibility odds (Fig. 1; Hood et al. 2015). We found here that the two mating-type loci in M. lagerheimii, although located on separate chromosomes, are completely linked to their respective centromeres, as in M. saponariae. Furthermore, we showed that in these sister species of anther-smut fungi such linkage occurred through independent recombination cessation events. Convergence of mating-type loci and centromere linkage seems to have also occurred at much larger phylogenetic scale within fungi. Cryptococcus amylolentus, a distant Microbotryum fungal relative, also displays both HD and PR genes linked to different centromeres (Sun et al. 2017a). Our study thus shows that natural selection can lead repeatedly to similar genomic changes but also to distinct and equally beneficial solutions under a shared evolutionary pressure. These findings contribute to our understanding of evolution and the degree to which it is repeatable.
Segregation analyses showed that the HD and PR loci are linked to their respective centromere in M. lagerheimii, as previously shown in its sister species M. saponariae (Hood et al. 2015). Although the inclusion of a finite number of analyzed meiotic tetrads leaves the possibility that linkage to centromeres is only nearly complete, our genomic results support complete linkage in the PR chromosome. We found substantial differentiation between alleles associated to alternative mating types at genes between the PR-proximal region and its centromere in both species, as well as a large inversion in M. saponariae. Additional evidence of complete recombination cessation is that the HD chromosome in M. saponariae and the PR chromosomes in both species are size dimorphic, with size cosegregating with mating-type alleles (Hood et al. 2015). Chromosome size dimorphism is likely due to the differential transposable element amounts we found between alternate HD and PR chromosomes in each species.
The absence of trans-specific polymorphism and the more recent inferred linkage dates compared to the speciation event between M. saponariae and M. lagerheimii strongly support that recombination cessation was independent in the two species. The recent origins of complete linkage between the two mating-type loci and their centromeres in both species are corroborated by the low synonymous divergence values between a1- and a2-associated alleles and the lack of extensive rearrangements in the regions without recombination between the mating-type locus proximal regions and the centromeres.
For linkage of mating-type loci to centromeres to be beneficial under selfing by automixis, both HD and PR mating-type genes need to be linked to their respective centromere (Zakharov 1986, 2005). However, for both M. lagerheimii and M. saponariae, the PR linkage to the centromere evolved long before the HD-centromere linkage. The PR locus-centromere linkage alone provides no advantage concerning gamete compatibility odds when mating occurs within a tetrad; however, the PR-centromere linkage may have been generated in several steps extending the recombination cessation region beyond mating-type genes, as previously described (Branco et al. 2017). This would be consistent with the apparent heterogeneity in the dS values in the genes between the PR-proximal region and the centromere, with highest dS values for genes closer to PR than to the centromere (Fig. 4). Under this hypothesis, expansion of the regions of suppressed recombination would have occurred through processes unrelated to the mating system, such as the accumulation and methylation of transposable elements in nonrecombining regions and in their margins (Ponnikas et al. 2018). Once the PR-centromere linkage was achieved, selection for HD-centromere linkage may have occurred and been selected for increasing odds of compatibility under automixis.
Alternatively, the close physical proximity of the HD locus to the centromere may be sufficient to render recombination events infrequent enough that recombination cessation between the PR locus and its centromere would be immediately beneficial for increasing the odds of gamete compatibility under automixis. This hypothesis of rare recombination between the centromere and the HD would explain the very low dS values in this region and the collinearity between b1 and b2 HD mating-type chromosomes in M. saponariae despite the inversion that occurred between HD and the centromere since its speciation from M. lagerheimii. This hypothesis is not incompatible with the PR-centromere linkage having evolved by successive evolutionary steps. Low recombination rates have been invoked in sex chromosomes to explain low differentiation between alleles on X and Y Chromosomes in some animals (Stöck et al. 2013).
Although inversions are often thought to play a major role in suppressing recombination (Lemaitre et al. 2009; Wang et al. 2012; Wright et al. 2016), nonrecombining regions with conserved collinearity have been reported in several fungi (Jacobson 2005; Grognet et al. 2014; Branco et al. 2017, 2018; Sun et al. 2017b). In this study, we only found inversions in the region with recent recombination cessation between the M. saponariae PR chromosomes. Finding that the limits of this inversion are precisely the centromere and the PR-proximal region is consistent with a role of inversions in recombination suppression, although we cannot exclude that the inversion occurred as a subsequent rearrangement after recombination cessation. The remaining mating-type loci and centromere linkage events occurred via different proximal mechanisms not involving rearrangements. Elucidating the proximal mechanisms suppressing recombination by exploring, for example, changes in DNA methylation and heterochromatin marks, as well as Spo11-dependent formation of double-strand breaks (Keeney 2008; Termolino et al. 2016), will be interesting in future studies.
Our results have general implications beyond mating systems in fungi and evolutionary convergence, providing an excellent illustration of the benefits of mating via central fusion automixis. Broader implication of central fusion automixis has been rarely considered, despite occurring in a wide range of taxa such as in fungi (Hood and Antonovics 2000; Zakharov 2005; Menkis et al. 2008; Grognet et al. 2014), plants (Asker 1980; Walker 1985; Antonius and Nybom 1995; Cruden and Lloyd 1995; Schön et al. 2009), and insects (Suomalainen et al. 1976; Normark 2003; Oldroyd et al. 2008). Theoretical models and reviews have highlighted the evolutionary and genetic consequences of central fusion automixis in maintaining heterozygosity (Zakharov 1986, 2005; Antonovics and Abrams 2004; Hood et al. 2005; Engelstädter 2017), but few cases have been experimentally studied so far. Our findings illustrate how linkage to centromere under central fusion automixis can generate a sort of pseudolinkage among genes on different chromosomes and preserve heterozygosity. Such maintenance of heterozygosity can be beneficial in a variety of cases (Ferreira and Amos 2006), such as the sheltering of deleterious alleles (Hood and Antonovics 2000) or overdominance in immune systems (Hraber et al. 2007) and other functions, as suggested at several loci in the case of the central fusion automictic Cape honeybees (Goudie et al. 2014).
Methods
To conduct segregation analyses, we isolated M. lagerheimii haploid cells from opposite poles of meiosis I across replicate meioses from the same diploid parent using micromanipulation (Hood et al. 2015). We investigated mating-type segregation by PCR amplification of allele-specific markers. The M. lagerheimii strain used for segregation analyses was collected on Lychnis flos-jovis in Valle Pesio, Italy (GPS 44.188400, 7.670650).
Genome analyses were conducted in the M. saponariae and M. lagerheimii assemblies. We used alternative mating types isolated from a single diploid spore of M. saponariae parasitizing Saponariae officinalis (cell 1268, PRAT 47, a1 b1, and cell 1269, PRAT 48, a2 b2) collected near Chiusa di Pesio, Italy (GPS coordinates 44.31713297, 7.622967437 on July 8, 2012). We used the M. lagerheimii genome previously published (a1 b1 and a2 b2 assemblies GCA_ 900015505.1 and GCA_900013405.1, respectively) (Branco et al. 2017). DNA was extracted with the QIAGEN Genomic-tip 100/G (catalog number 10243) and Genomic DNA Buffer Set (catalog number 19060) following the manufacturer's instructions and using a Carver hydraulic press (catalog number 3968). Haploid genomes were sequenced using the P6/C4 Pacific Biosciences SMRT technology (UCSD IGM Genomics Facility). Assemblies of the genomes were generated with the wgs-8.2 version of the PBcR assembler (Koren et al. 2012). Contigs were aligned with optical maps of the two mating-type chromosomes obtained previously (Hood et al. 2015), with MapSolver software (OpGen); see statistics on assemblies in Supplemental Tables S1 and S2. We obtained orthologous groups with orthAgogue (Ekseth et al. 2014) based on BLASTP + 2.2.30 followed by Markov clustering (Van Dongen 2000). We aligned the protein sequences of 780 fully conserved single-copy genes with MAFFT v7.388 (Katoh and Standley 2013) and obtained the codon-based CDS alignments with TranslatorX (Abascal et al. 2010). We used RAxML 8.2.7 (Stamatakis 2006) to obtain maximum likelihood gene trees for all 780 fully conserved single-copy genes and a species tree with the concatenated alignment. We estimated synonymous divergence (dS) and its standard error with the yn00 program of the PAML package (Yang 2007).
We used nine orthologous groups (8525 aligned codons) for dating the recombination cessation between the HD-proximal region and the centromere and 10 orthologous (10,200 aligned codons) groups for dating recombination cessation between the PR-proximal region and the centromere. Divergence times were estimated using BEAST v2.4.0 (Drummond and Rambaut 2007), with the XLM inputs being generated using BEAUTi (Drummond et al. 2012).
Transposable elements were identified and annotated de novo in the high-quality genome assemblies, using both LTR-harvest (defaults parameters) (Ellinghaus et al. 2008); and RepeatModeler (defaults parameters) (Smit and Hubley 2015). We identified de novo centromeric-specific repeats (Melters et al. 2013) using Tandem-Repeat Finder (TRF v. 4.07b) (Benson 1999); see Supplemental File 1.
Data access
The Pacific Biosciences (PacBio) genome assemblies from this study have been submitted to the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) under accession number GCA_900015975 for the a1 genome and GCA_900015475 for the a2 genome of Microbotryum saponariae from Saponaria officinalis.
Acknowledgments
This work was supported by the European Research Council starting grant GenomeFun 309403; National Science Foundation Division of Environmental Biology grant 1115765 and National Institutes of Health R15GM119092 grant to M.E.H.; the Marie Curie European grant 701646 and the Montana State University Agricultural Research Station to S.B.; and postdoctoral fellowship (SFRH/BPD/79198/2011) from Fundação para a Ciência e a Tecnologia, Portugal to M.A.C. We thank Cécile Fairhead for help with DNA extraction. PacBio sequencing was conducted at the IGM Genomics Center, University of California, San Diego, La Jolla, California.
Author contributions: T.G. and M.E.H. designed and supervised the study. T.G., M.E.H., and S.B. contributed to obtain funding. M.E.H., A.S., and T.G. obtained the genomes. F.C., R.C.R.D.L.V., S.B., and M.A.C. performed the genomic analyses. M.E.H. and A.S. performed the segregation analyses. F.C., T.G., and M.E.H. wrote the manuscript with contributions from all other authors.
Footnotes
-
↵5 These authors supervised the study.
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.242578.118.
- Received August 2, 2018.
- Accepted April 29, 2019.
This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date (see http://genome.cshlp.org/site/misc/terms.xhtml). After six months, it is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.