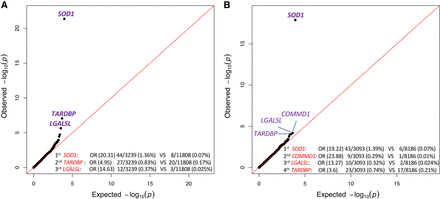
Intolerance-informed gene-level collapsing with unified/diversified ancestry samples. (A) A Q-Q plot presenting the results of the gene-based intolerance-informed collapsing of 3239 cases and 11,808 controls from diversified ancestries. Missense variants are aggregated only if they reside in an intolerant domain that is lower than the 50th percentile OE-ratio score, while loss-of-function variants are aggregated independent of location; 17,795 genes passed QC with more than one case or control carrier for this test. The genes with the top associations are labeled. λ = 1.14. (B) A Q-Q plot of a gene-based intolerance-informed collapsing of 3093 cases and 8186 controls of European ancestry; 18,135 genes passed QC with more than one case or control carrier for this test. The genes with the top associations are labeled and genome-wide significant genes are in bold. λ = 1.073.











