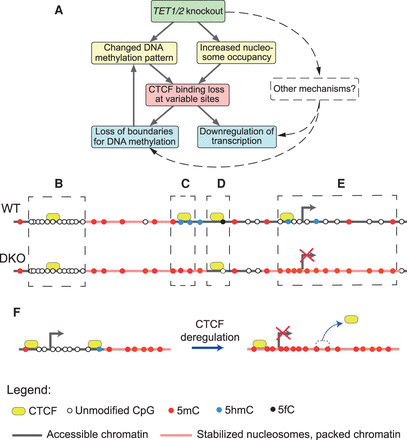
A scheme of different regimes of CTCF sensitivity to DNA modifications. (A) Simplified scheme of the possible causality of events: Tet1/2 knockout leads to the changed DNA methylation pattern and increased nucleosome occupancy. These lead to CTCF binding loss at variable sites. As a result, methylation spreads to larger areas, and neighboring genes are down-regulated. (B) Common CTCF sites were significantly enriched at CpG islands where DNA was unmethylated in both cell types, and CTCF binding was mostly determined by the DNA sequence. (C) CTCF sites marked by 5hmC in WT were predisposed for loss of CTCF binding in DKO cells, which could be accompanied by a 5hmC/5mC switch and the loss of 5hmC. (D) Regions near 5fC sites were more enriched for common than for lost CTCF sites. (E) CTCF loss at promoters and in the vicinity of genes may lead to the spreading of DNA methylation into neighboring regions as a function of the distance from the CTCF site. Genes inside such regions tend to become down-regulated in DKO ESCs. (F) In some cases, methylation of a single CpG inside a CTCF binding site may lead to CTCF removal, or vice versa, and results in the loss of the boundary between methylation microdomains. This process may induce a subsequent change of transcription, as shown for an example genomic region in Supplemental Figure S27.











