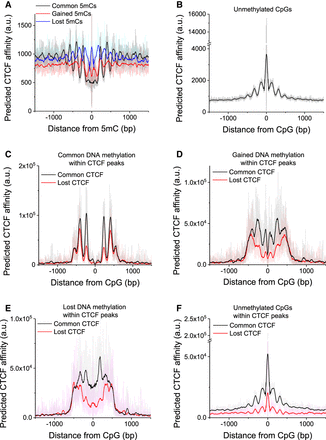
CTCF-DNA binding affinity predicted from the DNA sequence as a function of distance from CpGs. (A,B) Calculations performed for four classes of CpGs genome-wide. (A) CpGs that were commonly methylated in both cell states (methylation >0.8 both in WT and DKO cells; N = 10,439,081), that gained methylation (<0.2 in WT and >0.5 in DKO ESCs; N = 9,596,997), and that lost methylation (>0.5 in WT and <0.2 in DKO ESCs; N = 6,859,738). (B) Unmethylated CpGs (<0.2 in both WT and DKO; N = 15,316,892). (C–F) Calculations performed only for CpGs within CTCF ChIP-seq peaks in WT. (C) Common 5mC sites inside common (N = 25,740) and lost (N = 33,060) CTCF peaks. (D) Gained 5mC sites inside common (N = 37,702) and lost (N = 35,518) CTCF peaks. (E) Lost 5mC sites inside common (N = 35,632) and lost (N = 35,527) CTCF peaks. (F) Unmethylated CpGs inside common (N = 460,752) and lost (N = 179,288) CTCF peaks.











