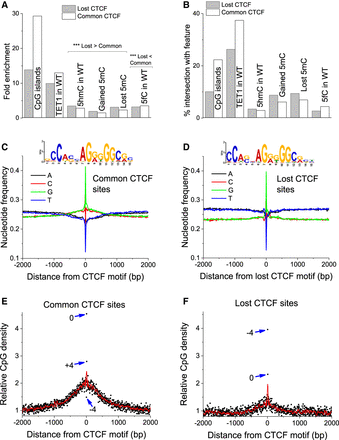
Loss of CTCF is associated with reduced GC content and CpG density. (A,B) Fold enrichment (A) and percentage overlap (B) of lost/common CTCF sites with different genomic features. CTCF sites are defined as 19-nucleotide motifs within the corresponding CTCF ChIP-seq peaks. (C,D) The nucleotide frequencies within ±2000 bp around CTCF motifs in common (C) and lost (D) peaks, as well as the corresponding consensus motifs. (E,F) CpG density around CTCF motifs in common and lost sites. Black dots correspond to individual CpG positions, red lines represent a spline interpolation of their density, and blue arrows indicate the outstanding CpGs inside the CTCF binding motif together with their coordinates with respect to the central peak of CpG density.











