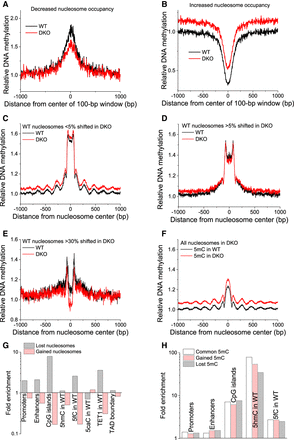
DNA methylation is associated with nucleosome repositioning. (A,B) Relative DNA methylation is shown around centers of 100-bp genomic regions with lost (A) and gained (B) nucleosome occupancy. (C–F) Changes in DNA methylation were associated with shifted nucleosomes. Relative DNA methylation is plotted as a function of the distance from the centers of nucleosomes on Chromosome 19 determined by paired-end MNase-assisted H3 ChIP-seq. Black lines indicate DNA methylation in WT; red lines, DNA methylation in DKO ESCs. Within each plot, WT and DKO methylation was normalized in the same way and is quantitatively comparable. (C) Common nucleosomes whose boundaries change <5% between WT and DKO ESCs (>95% overlap between the bodies of the corresponding paired-end reads in WT and DKO cells). (D) Nucleosomes in WT cells whose boundaries were changed in DKO by >5% (<95% overlap). (E) Nucleosomes in WT cells whose boundaries were changed in DKO by >30% (<70% overlap). (F) All nucleosomes in DKO ESCs. (G) Fold enrichment of lost/gained nucleosomes at different genomic features. (H) Fold enrichment of common/gained/lost 5mC at genomic features.











