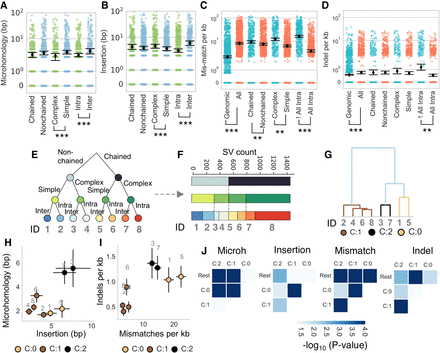
Differential end processing reveals distinct pathways used in repair of chained SVs. (A,B) Differences in microhomology and insertion profiles of SVs were quantified according to a classification of SVs as chained versus nonchained, complex versus simple, and intrachromosomal versus interchromosomal. (C,D) Upstream and downstream sequences were also analyzed for the numbers of mismatches and indels, with the All Intra versus !All Intra comparison referring to contigs with entirely intrachromosomal events versus contigs with one or more translocation. SVs were binned into eight groups (E) with the numbers of each division of the data presented in F with the category ID numbered in gray. (G) Agglomerative clustering was used to identify groups with similar properties labeled from C:0 to C:2. The mean values for microhomology and insertion (H), and mismatches and indels (I) of these groups are visualized as scatterplots in H and I with the color relating to the cluster in panel G and the gray text annotation referring to the category ID from panel E. (J) A pairwise statistical test was used to compare features and identify significant differences between clusters. (**) P-value <0.005; (***) P-value <0.0005; Mann–Whitney U test. Error bars, SEM.











