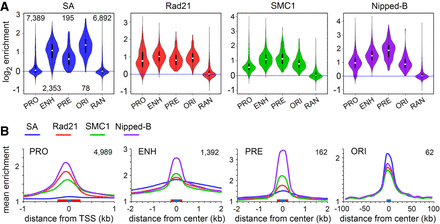
Cohesin subunits are present in different ratios at promoters, enhancers, PREs, and replication origins in BG3 cells. (A) Violin plot distributions of SA (blue), Rad21 (red), SMC1 (green), and Nipped-B (purple) at active promoters (PRO), enhancers (ENH), Polycomb Response Elements (PRE), early replication origin centers (ORI), and randomly positioned sequences (RAN). The numbers of each type of feature analyzed are indicated in the SA plot. White dots are the median values given in Supplemental Table S2. (B) Meta-analyses of promoters, enhancers, PREs, and early replication origins for SA (blue), Rad21 (red), SMC1 (green), and Nipped-B (purple). Red boxes on the x-axes indicate the feature sizes used to calculate occupancy for the violin plots, and blue boxes indicate the bin sizes used to average enrichment for the meta-analysis. The numbers of each type of feature used for meta-analysis are indicated in the upper right corner of each graph. These are less than for the violin plots because features that overlap in the meta-analysis region were removed to minimize distortions.











